Science
-
 Deep Learning Predicts Drug-Drug and Drug-Food Interactions
A Korean research team from KAIST developed a computational framework, DeepDDI, that accurately predicts and generates 86 types of drug-drug and drug-food interactions as outputs of human-readable sentences, which allows in-depth understanding of the drug-drug and drug-food interactions.
Drug interactions, including drug-drug interactions (DDIs) and drug-food constituent interactions (DFIs), can trigger unexpected pharmacological effects, including adverse drug events (ADEs), with causal mechanisms often unknown. However, current prediction methods do not provide sufficient details beyond the chance of DDI occurrence, or require detailed drug information often unavailable for DDI prediction.
To tackle this problem, Dr. Jae Yong Ryu, Assistant Professor Hyun Uk Kim and Distinguished Professor Sang Yup Lee, all from the Department of Chemical and Biomolecular Engineering at Korea Advanced Institute of Science and Technology (KAIST), developed a computational framework, named DeepDDI, that accurately predicts 86 DDI types for a given drug pair. The research results were published online in Proceedings of the National Academy of Sciences of the United States of America (PNAS) on April 16, 2018, which is entitled “Deep learning improves prediction of drug-drug and drug-food interactions.”
DeepDDI takes structural information and names of two drugs in pair as inputs, and predicts relevant DDI types for the input drug pair. DeepDDI uses deep neural network to predict 86 DDI types with a mean accuracy of 92.4% using the DrugBank gold standard DDI dataset covering 192,284 DDIs contributed by 191,878 drug pairs. Very importantly, DDI types predicted by DeepDDI are generated in the form of human-readable sentences as outputs, which describe changes in pharmacological effects and/or the risk of ADEs as a result of the interaction between two drugs in pair. For example, DeepDDI output sentences describing potential interactions between oxycodone (opioid pain medication) and atazanavir (antiretroviral medication) were generated as follows: “The metabolism of Oxycodone can be decreased when combined with Atazanavir”; and “The risk or severity of adverse effects can be increased when Oxycodone is combined with Atazanavir”. By doing this, DeepDDI can provide more specific information on drug interactions beyond the occurrence chance of DDIs or ADEs typically reported to date.
DeepDDI was first used to predict DDI types of 2,329,561 drug pairs from all possible combinations of 2,159 approved drugs, from which DDI types of 487,632 drug pairs were newly predicted. Also, DeepDDI can be used to suggest which drug or food to avoid during medication in order to minimize the chance of adverse drug events or optimize the drug efficacy. To this end, DeepDDI was used to suggest potential causal mechanisms for the reported ADEs of 9,284 drug pairs, and also predict alternative drug candidates for 62,707 drug pairs having negative health effects to keep only the beneficial effects. Furthermore, DeepDDI was applied to 3,288,157 drug-food constituent pairs (2,159 approved drugs and 1,523 well-characterized food constituents) to predict DFIs. The effects of 256 food constituents on pharmacological effects of interacting drugs and bioactivities of 149 food constituents were also finally predicted. All these prediction results can be useful if an individual is taking medications for a specific (chronic) disease such as hypertension or diabetes mellitus type 2.
Distinguished Professor Sang Yup Lee said, “We have developed a platform technology DeepDDI that will allow precision medicine in the era of Fourth Industrial Revolution. DeepDDI can serve to provide important information on drug prescription and dietary suggestions while taking certain drugs to maximize health benefits and ultimately help maintain a healthy life in this aging society.”
Figure 1. Overall scheme of Deep DDDI and prediction of food constituents that reduce the in vivo concentration of approved drugs
2018.04.18 View 13872
Deep Learning Predicts Drug-Drug and Drug-Food Interactions
A Korean research team from KAIST developed a computational framework, DeepDDI, that accurately predicts and generates 86 types of drug-drug and drug-food interactions as outputs of human-readable sentences, which allows in-depth understanding of the drug-drug and drug-food interactions.
Drug interactions, including drug-drug interactions (DDIs) and drug-food constituent interactions (DFIs), can trigger unexpected pharmacological effects, including adverse drug events (ADEs), with causal mechanisms often unknown. However, current prediction methods do not provide sufficient details beyond the chance of DDI occurrence, or require detailed drug information often unavailable for DDI prediction.
To tackle this problem, Dr. Jae Yong Ryu, Assistant Professor Hyun Uk Kim and Distinguished Professor Sang Yup Lee, all from the Department of Chemical and Biomolecular Engineering at Korea Advanced Institute of Science and Technology (KAIST), developed a computational framework, named DeepDDI, that accurately predicts 86 DDI types for a given drug pair. The research results were published online in Proceedings of the National Academy of Sciences of the United States of America (PNAS) on April 16, 2018, which is entitled “Deep learning improves prediction of drug-drug and drug-food interactions.”
DeepDDI takes structural information and names of two drugs in pair as inputs, and predicts relevant DDI types for the input drug pair. DeepDDI uses deep neural network to predict 86 DDI types with a mean accuracy of 92.4% using the DrugBank gold standard DDI dataset covering 192,284 DDIs contributed by 191,878 drug pairs. Very importantly, DDI types predicted by DeepDDI are generated in the form of human-readable sentences as outputs, which describe changes in pharmacological effects and/or the risk of ADEs as a result of the interaction between two drugs in pair. For example, DeepDDI output sentences describing potential interactions between oxycodone (opioid pain medication) and atazanavir (antiretroviral medication) were generated as follows: “The metabolism of Oxycodone can be decreased when combined with Atazanavir”; and “The risk or severity of adverse effects can be increased when Oxycodone is combined with Atazanavir”. By doing this, DeepDDI can provide more specific information on drug interactions beyond the occurrence chance of DDIs or ADEs typically reported to date.
DeepDDI was first used to predict DDI types of 2,329,561 drug pairs from all possible combinations of 2,159 approved drugs, from which DDI types of 487,632 drug pairs were newly predicted. Also, DeepDDI can be used to suggest which drug or food to avoid during medication in order to minimize the chance of adverse drug events or optimize the drug efficacy. To this end, DeepDDI was used to suggest potential causal mechanisms for the reported ADEs of 9,284 drug pairs, and also predict alternative drug candidates for 62,707 drug pairs having negative health effects to keep only the beneficial effects. Furthermore, DeepDDI was applied to 3,288,157 drug-food constituent pairs (2,159 approved drugs and 1,523 well-characterized food constituents) to predict DFIs. The effects of 256 food constituents on pharmacological effects of interacting drugs and bioactivities of 149 food constituents were also finally predicted. All these prediction results can be useful if an individual is taking medications for a specific (chronic) disease such as hypertension or diabetes mellitus type 2.
Distinguished Professor Sang Yup Lee said, “We have developed a platform technology DeepDDI that will allow precision medicine in the era of Fourth Industrial Revolution. DeepDDI can serve to provide important information on drug prescription and dietary suggestions while taking certain drugs to maximize health benefits and ultimately help maintain a healthy life in this aging society.”
Figure 1. Overall scheme of Deep DDDI and prediction of food constituents that reduce the in vivo concentration of approved drugs
2018.04.18 View 13872 -
 Two Professors Receive the Asan Medical Award
(Professor Ho Min Kim and Chair Profesor Eunjoon Kim (from far right)
Chair Professor Eunjoon Kim of the Department of Biological Sciences and Professor Ho Min Kim from the Graduate School of Medical Science & Engineering won the 11th Asan Medical Award in the areas of basic medicine and young medical scholar on March 21.
The Asan Medical Award has been recognizing the most distinguished scholars in the areas of basic and clinical medicines annually since 2007.
Chair Professor Kim won the 300 million KRW award in recognition of his research in the mechanism of synaptic brain dysfunction and its relation with neural diseases.
The young medical scholar’s award recognizes a promising scholar under the age of 40. Professor Kim won the award for identifying the key protein structure and molecular mechanism controlling immunocytes and neurons. He earned a 50 million KRW prize.
2018.03.26 View 11833
Two Professors Receive the Asan Medical Award
(Professor Ho Min Kim and Chair Profesor Eunjoon Kim (from far right)
Chair Professor Eunjoon Kim of the Department of Biological Sciences and Professor Ho Min Kim from the Graduate School of Medical Science & Engineering won the 11th Asan Medical Award in the areas of basic medicine and young medical scholar on March 21.
The Asan Medical Award has been recognizing the most distinguished scholars in the areas of basic and clinical medicines annually since 2007.
Chair Professor Kim won the 300 million KRW award in recognition of his research in the mechanism of synaptic brain dysfunction and its relation with neural diseases.
The young medical scholar’s award recognizes a promising scholar under the age of 40. Professor Kim won the award for identifying the key protein structure and molecular mechanism controlling immunocytes and neurons. He earned a 50 million KRW prize.
2018.03.26 View 11833 -
 Scientist of March, Professor Hee-Seung Lee
(Professor Hee-Seung Lee)
Professor Hee-Seung Lee from the Department of Chemistry at KAIST received the ‘Science and Technology Award of the Month’ awarded by the Ministry of ICT and Science, and the National Research Foundation of Korea for March 2018.
Professor Lee has been recognized for successfully producing peptide-based molecular machines, which used to be made of metals.
The methodology can be translated into magnetotactic behavior at the macroscopic scale, which is reminiscent of magnetosomes in magnetotactic bacteria.
The team employed foldectures, self-assembled molecular architectures of β-peptide foldamers, to develop the peptide-based molecular machines that uniformly align with respect to an applied static magnetic field.
Professor Lee said, “Molecular machines are widely used in the field of medical engineering or material science; however, there were limitations for developing the machines using magnetic fields. By developing peptide-based molecular machines, we were able to develop body-friendly molecular machines.”
Every month, the Ministry of ICT and Science and the National Research Foundation of Korea award a cash prize worth 10,000,000 KRW to a scientist who has contributed to science and technology with outstanding research and development performance.
2018.03.15 View 11658
Scientist of March, Professor Hee-Seung Lee
(Professor Hee-Seung Lee)
Professor Hee-Seung Lee from the Department of Chemistry at KAIST received the ‘Science and Technology Award of the Month’ awarded by the Ministry of ICT and Science, and the National Research Foundation of Korea for March 2018.
Professor Lee has been recognized for successfully producing peptide-based molecular machines, which used to be made of metals.
The methodology can be translated into magnetotactic behavior at the macroscopic scale, which is reminiscent of magnetosomes in magnetotactic bacteria.
The team employed foldectures, self-assembled molecular architectures of β-peptide foldamers, to develop the peptide-based molecular machines that uniformly align with respect to an applied static magnetic field.
Professor Lee said, “Molecular machines are widely used in the field of medical engineering or material science; however, there were limitations for developing the machines using magnetic fields. By developing peptide-based molecular machines, we were able to develop body-friendly molecular machines.”
Every month, the Ministry of ICT and Science and the National Research Foundation of Korea award a cash prize worth 10,000,000 KRW to a scientist who has contributed to science and technology with outstanding research and development performance.
2018.03.15 View 11658 -
 KAIST Professors Selected as Y-KAST Members
Professor YongKeun Park, Professor Bumjoon Kim, Professor Keon Jae Lee, and Professor Young Seok Ju were selected as the newest members of the Young Korean Academy of Science and Technology (Y-KAST).
The Korean Academy of Science and Technology, an academic institution of professional experts, selected 26 promising scientists under the age of 43 to join Y-KAST. and four KAIST professors were included in the list.
The newest members were conferred on February 26.
Research Field
Name
Natural Sciences
YongKeun Park (Dept. of Physics)
Engineering
Bumjoon Kim (Dept. of Chemical and Biomolecular Engineering)
Agricultural & Fishery Sciences
Keon Jae Lee (Dept. of Materials Science and Engineering)
Medical Sciences
Young Seok Ju (Graduate School of Medical Science and Engineering)
2018.03.05 View 11384
KAIST Professors Selected as Y-KAST Members
Professor YongKeun Park, Professor Bumjoon Kim, Professor Keon Jae Lee, and Professor Young Seok Ju were selected as the newest members of the Young Korean Academy of Science and Technology (Y-KAST).
The Korean Academy of Science and Technology, an academic institution of professional experts, selected 26 promising scientists under the age of 43 to join Y-KAST. and four KAIST professors were included in the list.
The newest members were conferred on February 26.
Research Field
Name
Natural Sciences
YongKeun Park (Dept. of Physics)
Engineering
Bumjoon Kim (Dept. of Chemical and Biomolecular Engineering)
Agricultural & Fishery Sciences
Keon Jae Lee (Dept. of Materials Science and Engineering)
Medical Sciences
Young Seok Ju (Graduate School of Medical Science and Engineering)
2018.03.05 View 11384 -
 Successful Synthesis of Gamma-Lanctam Rings from Hydrocarbons
(The team of Professor Chang, far right, at the Department of Chemistry)
KAIST chemists have designed a novel strategy to synthesize ring-shaped cyclic molecules, highly sought-after by pharmaceutical and chemical industries, and known as gamma-lactams. This study describes how these five-membered rings can be prepared from inexpensive and readily available feedstock hydrocarbons, as well as from complex organic molecules, such as amino acids and steroids.
Gamma-lactams find several applications in medicinal, synthetic, and material chemistry. For example, they are included in a large number of pharmaceutically active compounds with antibiotic, anti-inflammatory, and anti-tumoral functions. This research was published in Science on March 2.
Conversion of hydrocarbons into nitrogen-containing compounds is an important area of research, where the challenge lies in breaking strong carbon-hydrogen (C−H) bonds, and converting them into carbon-nitrogen (C–N) bonds in a controlled fashion. For this reason, hydrocarbons are difficult to use as starting materials, albeit the fact that they exist in large quantities in nature.
Over the last 35 years, chemists have found ways of converting simple hydrocarbons into nitrogen-containing rings, such as indoles or pyrrolidines, but gamma-lactams proved impossible to prepare using the same approaches. Researchers hypothesized that such failure was due to alternative chemical pathways that steer the reaction away from the wanted rings: The reaction intermediate (carbonylnitrene) quickly breaks down into unsought products. Using computer models of the desired and undesired reaction pathways, the team found a strategy to completely shut down the latter in order to obtain the longed-for gamma-lactams. For the first time, these four carbons and one nitrogen cyclic molecules were obtained directly from simple feedstock chemicals.
Led by Professor Chang Sukbok at the Department of Chemistry, the team designed the winning reaction with the help of computer simulations that analyze the reaction mechanisms and calculate the energy required for the reaction to take place. According to such computer predictions, the reaction could follow three pathways, leading to the formation of either the desired gamma-lactam, an unwanted product (isocyanate), or the degradation of the catalyst caused by the substrate reacting with the catalyst backbone. Combining experimental observations and detailed computer simulations, the team designed an iridium-based catalyst, highly selective for the gamma-lactam formation. In this way, the two undesired pathways were systematically shut down, leaving the formation of the nitrogen-containing ring as the only possible outcome. Professor Chang is also in charge of the Center for Catalytic Hydrocarbon Functionalizations at the Institute for Basic Science (IBS).
“With this work we offer a brand new solution to a long-standing challenge and demonstrate the power of what we call mechanism-based reaction development,” explains Professor Baik Mu-Hyun, a corresponding author of the study.
Beyond using cheap feedstock hydrocarbons as substrates, the team was also successful in converting amino acids, steroids, and other bio-relevant molecules into gamma-lactams, which might find a variety of applications as plant insecticide, drugs against parasitic worms, or anti-aging agents. This new synthetic technology gives much easier access to these complicated molecules and will enable the development of potential drugs in a much shorter amount of time at a lower cost.
Figure 1: Selective amidation reaction using newly designed iridium (Ir) catalysts. Abundant in nature Hydrocarbons are used as substrates to synthesize nitrogen-containing ring, called gamma-lactams.
Figure 2: Three possible reaction pathways and energy barriers predicted by computational chemistry. The scientists developed new iridium-based catalysts that are highly selective for the C–H insertion pathway which leads to the desired gamma-lactam molecules.
Figure 3: Interesting gamma-lactams derived from natural and unnatural amino acids, steroids, etc., which may be used to protect plants against insects, fight parasitic worms, or as anti-aging agents.
2018.03.02 View 9751
Successful Synthesis of Gamma-Lanctam Rings from Hydrocarbons
(The team of Professor Chang, far right, at the Department of Chemistry)
KAIST chemists have designed a novel strategy to synthesize ring-shaped cyclic molecules, highly sought-after by pharmaceutical and chemical industries, and known as gamma-lactams. This study describes how these five-membered rings can be prepared from inexpensive and readily available feedstock hydrocarbons, as well as from complex organic molecules, such as amino acids and steroids.
Gamma-lactams find several applications in medicinal, synthetic, and material chemistry. For example, they are included in a large number of pharmaceutically active compounds with antibiotic, anti-inflammatory, and anti-tumoral functions. This research was published in Science on March 2.
Conversion of hydrocarbons into nitrogen-containing compounds is an important area of research, where the challenge lies in breaking strong carbon-hydrogen (C−H) bonds, and converting them into carbon-nitrogen (C–N) bonds in a controlled fashion. For this reason, hydrocarbons are difficult to use as starting materials, albeit the fact that they exist in large quantities in nature.
Over the last 35 years, chemists have found ways of converting simple hydrocarbons into nitrogen-containing rings, such as indoles or pyrrolidines, but gamma-lactams proved impossible to prepare using the same approaches. Researchers hypothesized that such failure was due to alternative chemical pathways that steer the reaction away from the wanted rings: The reaction intermediate (carbonylnitrene) quickly breaks down into unsought products. Using computer models of the desired and undesired reaction pathways, the team found a strategy to completely shut down the latter in order to obtain the longed-for gamma-lactams. For the first time, these four carbons and one nitrogen cyclic molecules were obtained directly from simple feedstock chemicals.
Led by Professor Chang Sukbok at the Department of Chemistry, the team designed the winning reaction with the help of computer simulations that analyze the reaction mechanisms and calculate the energy required for the reaction to take place. According to such computer predictions, the reaction could follow three pathways, leading to the formation of either the desired gamma-lactam, an unwanted product (isocyanate), or the degradation of the catalyst caused by the substrate reacting with the catalyst backbone. Combining experimental observations and detailed computer simulations, the team designed an iridium-based catalyst, highly selective for the gamma-lactam formation. In this way, the two undesired pathways were systematically shut down, leaving the formation of the nitrogen-containing ring as the only possible outcome. Professor Chang is also in charge of the Center for Catalytic Hydrocarbon Functionalizations at the Institute for Basic Science (IBS).
“With this work we offer a brand new solution to a long-standing challenge and demonstrate the power of what we call mechanism-based reaction development,” explains Professor Baik Mu-Hyun, a corresponding author of the study.
Beyond using cheap feedstock hydrocarbons as substrates, the team was also successful in converting amino acids, steroids, and other bio-relevant molecules into gamma-lactams, which might find a variety of applications as plant insecticide, drugs against parasitic worms, or anti-aging agents. This new synthetic technology gives much easier access to these complicated molecules and will enable the development of potential drugs in a much shorter amount of time at a lower cost.
Figure 1: Selective amidation reaction using newly designed iridium (Ir) catalysts. Abundant in nature Hydrocarbons are used as substrates to synthesize nitrogen-containing ring, called gamma-lactams.
Figure 2: Three possible reaction pathways and energy barriers predicted by computational chemistry. The scientists developed new iridium-based catalysts that are highly selective for the C–H insertion pathway which leads to the desired gamma-lactam molecules.
Figure 3: Interesting gamma-lactams derived from natural and unnatural amino acids, steroids, etc., which may be used to protect plants against insects, fight parasitic worms, or as anti-aging agents.
2018.03.02 View 9751 -
 The 2018 Commencement of KAIST at a Glance
KAIST awarded a total of 2, 736 degrees at the 2018 commencement ceremony on February 23. Among the honorees, Chairman and CEO of Samsung Electronics and Samsung Advanced Institute of Technology (SAIT) Oh-Hyun Kwon was recognized as the first alumnus honorary doctorate recipient of KAIST.
More than 5,000 family, friends, and graduates including distinguished guests of Minister of Science and ICT Young-Min Yu, the Member of National Assembly Kyung-Jin Kim, Chairman of the KAIST Board of Trustees Jang-Moo Lee, and the Chairperson of the KAIST Development Foundation Soo-Young Lee attended to celebrate the graduates. During the commencement, a total of 2,736 students earned degrees: 644 PhD degrees, 1,352 master’s degrees, and 740 bachelor’s degrees.
(Minister of Science and ICT Young-Min Yu)
(The Member of National Assembly Kyung-Jin Kim)
This year, Chairman and CEO of Samsung Electronics and SAIT Kwon shared the spotlight with many other graduates. Kwon received his Master’s degree in Electrical Engineering from KAIST in 1977 and completed his Ph.D. in Electrical Engineering from Stanford University in 1985. During his more than 33-year career at Samsung, he has made significant contribution to the development of 4M DRAM and the world’s first 64M DRAM. The success of 4M DRAM and 64 DRAM led Samsung to clinch the top position in the DRAM and NAND flash business around the world. This helped Samsung emerge as a global leader in the semiconductor industry.
(From left: Chairman and CEO of Samsung Electronics and SAIT Oh-Hyun Kwon and KAIST President Sung-Chul Shin)
During the commencement speech, Kwon and President Shin both highlighted the importance of collaboration instead of competition.
Kwon encouraged the graduates to understand others to make wonderful synergy. “When you first notice the true value of another person and interact with them, the value of the individual will be doubled and will bring about a greater impact,” he said.
Also, he stressed having a collaborative mindset by saying, “All of you here, including myself, are people who have benefited from society. We must cooperate with each other and give back to society for the vest results.”
While highlighting the core values of KAIST, creativity, challenge and caring, President Shin also emphasized collaboration with others. He said, “In the future, expertise in a single discipline will not lead to new inventions or discoveries. This highlights the importance of multidisciplinary, convergence research. The key to success lies in the acknowledgement of your peers as partners for mutual growth. Your partners will make up your weak areas and become your most important asset. May you expand your personal network by finding valuable partners not only within your laboratory and workplace, but beyond Korea.”
“Go out into the world and change it as a global shaper, global innovator, and global mover. I hope that each and every one of you will add benefits the world and your legacy will be remembered for generations to come. This is your obligation as a graduate of KAIST,” he said.
Click here to view the full text of President Sung-Chul Shin’s address to the graduates
+ List of academically outstanding undergraduate degree recipients who received honors during the Commencement 2018 of KAIST
Award
Department
Winner
Minister of Science and ICT Award
Dept. of Mathematical Sciences
Seong-Hyeok Park
KAIST Board Chairperson Award
School of Computing
Hyeong-Seok Kim
KAIST President Award
Dept. of Chemistry
Hoi-Min Cheong
KAIST Development Foundation Chairperson Award
Dept. of Biological Sciences
Gi-Song Kim
Dept. of Industrial & Systems Engineering
Seung-Hun Lee
2018.02.23 View 13771
The 2018 Commencement of KAIST at a Glance
KAIST awarded a total of 2, 736 degrees at the 2018 commencement ceremony on February 23. Among the honorees, Chairman and CEO of Samsung Electronics and Samsung Advanced Institute of Technology (SAIT) Oh-Hyun Kwon was recognized as the first alumnus honorary doctorate recipient of KAIST.
More than 5,000 family, friends, and graduates including distinguished guests of Minister of Science and ICT Young-Min Yu, the Member of National Assembly Kyung-Jin Kim, Chairman of the KAIST Board of Trustees Jang-Moo Lee, and the Chairperson of the KAIST Development Foundation Soo-Young Lee attended to celebrate the graduates. During the commencement, a total of 2,736 students earned degrees: 644 PhD degrees, 1,352 master’s degrees, and 740 bachelor’s degrees.
(Minister of Science and ICT Young-Min Yu)
(The Member of National Assembly Kyung-Jin Kim)
This year, Chairman and CEO of Samsung Electronics and SAIT Kwon shared the spotlight with many other graduates. Kwon received his Master’s degree in Electrical Engineering from KAIST in 1977 and completed his Ph.D. in Electrical Engineering from Stanford University in 1985. During his more than 33-year career at Samsung, he has made significant contribution to the development of 4M DRAM and the world’s first 64M DRAM. The success of 4M DRAM and 64 DRAM led Samsung to clinch the top position in the DRAM and NAND flash business around the world. This helped Samsung emerge as a global leader in the semiconductor industry.
(From left: Chairman and CEO of Samsung Electronics and SAIT Oh-Hyun Kwon and KAIST President Sung-Chul Shin)
During the commencement speech, Kwon and President Shin both highlighted the importance of collaboration instead of competition.
Kwon encouraged the graduates to understand others to make wonderful synergy. “When you first notice the true value of another person and interact with them, the value of the individual will be doubled and will bring about a greater impact,” he said.
Also, he stressed having a collaborative mindset by saying, “All of you here, including myself, are people who have benefited from society. We must cooperate with each other and give back to society for the vest results.”
While highlighting the core values of KAIST, creativity, challenge and caring, President Shin also emphasized collaboration with others. He said, “In the future, expertise in a single discipline will not lead to new inventions or discoveries. This highlights the importance of multidisciplinary, convergence research. The key to success lies in the acknowledgement of your peers as partners for mutual growth. Your partners will make up your weak areas and become your most important asset. May you expand your personal network by finding valuable partners not only within your laboratory and workplace, but beyond Korea.”
“Go out into the world and change it as a global shaper, global innovator, and global mover. I hope that each and every one of you will add benefits the world and your legacy will be remembered for generations to come. This is your obligation as a graduate of KAIST,” he said.
Click here to view the full text of President Sung-Chul Shin’s address to the graduates
+ List of academically outstanding undergraduate degree recipients who received honors during the Commencement 2018 of KAIST
Award
Department
Winner
Minister of Science and ICT Award
Dept. of Mathematical Sciences
Seong-Hyeok Park
KAIST Board Chairperson Award
School of Computing
Hyeong-Seok Kim
KAIST President Award
Dept. of Chemistry
Hoi-Min Cheong
KAIST Development Foundation Chairperson Award
Dept. of Biological Sciences
Gi-Song Kim
Dept. of Industrial & Systems Engineering
Seung-Hun Lee
2018.02.23 View 13771 -
 KAIST, First to Win the Cube Satellite Competition
Professor Hyochoong Bang from the Department of Aerospace Engineering and his team received the Minister of Science and ICT Award at the 1st Cube Satellite Competition.
The team actually participated in the competition in 2012, but it took several years for the awarding ceremony since it took years for the satellites to be designed, produced, and launched.
The KAIST team successfully developed a cube satellite, named ‘Little Intelligent Nanosatellite of KAIST (LINK)’ and completed its launch in April 2017.
LINK (size: 20cmx10cmx10cm, weight: 2kg) mounted mass spectrometry and Langmuir probe for Earth observation. The Langmuir probe was developed by Professor Kyoung Wook Min from the Department of Physics, KAIST.
Yeerang Lim, a PhD student from the Department of Aerospace Engineering said, “I still remember the feeling that I had on the day when LINK launched into orbit and sent back signals. I hope that space exploration is not something far away but attainable for us in near future.”
2018.02.22 View 14126
KAIST, First to Win the Cube Satellite Competition
Professor Hyochoong Bang from the Department of Aerospace Engineering and his team received the Minister of Science and ICT Award at the 1st Cube Satellite Competition.
The team actually participated in the competition in 2012, but it took several years for the awarding ceremony since it took years for the satellites to be designed, produced, and launched.
The KAIST team successfully developed a cube satellite, named ‘Little Intelligent Nanosatellite of KAIST (LINK)’ and completed its launch in April 2017.
LINK (size: 20cmx10cmx10cm, weight: 2kg) mounted mass spectrometry and Langmuir probe for Earth observation. The Langmuir probe was developed by Professor Kyoung Wook Min from the Department of Physics, KAIST.
Yeerang Lim, a PhD student from the Department of Aerospace Engineering said, “I still remember the feeling that I had on the day when LINK launched into orbit and sent back signals. I hope that space exploration is not something far away but attainable for us in near future.”
2018.02.22 View 14126 -
 Professor Il-Doo Kim Recevies the Song-gok Award
Professor Il-Doo Kim from the Department of Materials Science and Engineering at KAIST received the 20th Song-gok Science and Technology Award from Korea Institute of Science and Technology (KSIT).
The Song-gok Science and Technology Award was established to praise the accomplishments of the first president, Hyung-seop Choi, whose penname is Song-gok. The award selects a recipient in the field of materials and technology every other year.
Professor Kim, in recognition of his outstanding research and contributions to materials science in Korea, received the award during the 52nd anniversary ceremony of KIST on February 9.
Professor Kim focuses on developing nanofiber gas sensors for diagnosing disease in advance by analyzing exhaled biomarkers with electrospinning technology.
He has published more than 211 papers and has recorded more than 9,650 citations and 50 h-index.
Professor Kim has registered 107 patents and applied 38 patents in Korea while registering 29 patents and applying 16 patents overseas. Also, he transferred four technologies in 2017.
Professor Kim is recognized as one of the researchers who is leading nanofiber technology. On January 17, he made a keynote speech at the 5th International Conference on Electrospinning, which was his fourth keynote speech at that conference.
Moreover, he received the Technology Innovation Award at the College of Engineering, KAIST on December 19, 2017.
Professor Kim said, “It is my great honor to receive the Song-gok Science and Technology Award. I would like to bring distinction to KAIST by taking the lead in the commercializing a nanofiber-based highly sensitive nanosensors, diversifying and commercializing technology using nanofiber.”
2018.02.13 View 10380
Professor Il-Doo Kim Recevies the Song-gok Award
Professor Il-Doo Kim from the Department of Materials Science and Engineering at KAIST received the 20th Song-gok Science and Technology Award from Korea Institute of Science and Technology (KSIT).
The Song-gok Science and Technology Award was established to praise the accomplishments of the first president, Hyung-seop Choi, whose penname is Song-gok. The award selects a recipient in the field of materials and technology every other year.
Professor Kim, in recognition of his outstanding research and contributions to materials science in Korea, received the award during the 52nd anniversary ceremony of KIST on February 9.
Professor Kim focuses on developing nanofiber gas sensors for diagnosing disease in advance by analyzing exhaled biomarkers with electrospinning technology.
He has published more than 211 papers and has recorded more than 9,650 citations and 50 h-index.
Professor Kim has registered 107 patents and applied 38 patents in Korea while registering 29 patents and applying 16 patents overseas. Also, he transferred four technologies in 2017.
Professor Kim is recognized as one of the researchers who is leading nanofiber technology. On January 17, he made a keynote speech at the 5th International Conference on Electrospinning, which was his fourth keynote speech at that conference.
Moreover, he received the Technology Innovation Award at the College of Engineering, KAIST on December 19, 2017.
Professor Kim said, “It is my great honor to receive the Song-gok Science and Technology Award. I would like to bring distinction to KAIST by taking the lead in the commercializing a nanofiber-based highly sensitive nanosensors, diversifying and commercializing technology using nanofiber.”
2018.02.13 View 10380 -
 First Female Grand Prize Awardee of Samsung Humantech
Yeunhee Huh, PhD candidate (Professor Gyu-Hyeong Cho) from the School of Electrical Engineering received the grand prize of the 24th Humantech Paper Award. She is the first female recipient of this prize since its establishment in 1994. The Humantech Paper Award is hosted by Samsung Electronics and sponsored by the Ministry of Science and ICT with JoongAng Daily Newspaper. Her paper is titled, ‘A Hybrid Structure Dual-Path Step-Down Converter with 96.2% Peak Efficiency using 250mΩ Large-DCR Inductor’. Electronic devices require numerous chips and have a power converter to supply energy adequately. She proposed a new structure to enhance energy efficiency by combining inductors and capacitors. Enhancing energy efficiency can reduce energy loss, which prolongs battery hours and solves overheating of devices; for instance, energy loss leads to the overheating issue affecting phone chargers. This technology can be applied to various electronic devices, such as cell phones, laptops, and drones. Huh said, “Power has to go up in order to meet customers’ needs; however the overheating problem emerges during this process. This problem affects surrounding circuits and causes other issues, such as malfunctions of electronic devices. This technology may vary according to the conditions, but it can enhance energy efficiency up to 4%.”During the ceremony, about eight hundred million KRW worth cash prizes was conferred to 119 papers. KAIST (44 papers) and Gyeonggi Science High School (6 papers) received special awards given to the schools.
2018.02.12 View 11690
First Female Grand Prize Awardee of Samsung Humantech
Yeunhee Huh, PhD candidate (Professor Gyu-Hyeong Cho) from the School of Electrical Engineering received the grand prize of the 24th Humantech Paper Award. She is the first female recipient of this prize since its establishment in 1994. The Humantech Paper Award is hosted by Samsung Electronics and sponsored by the Ministry of Science and ICT with JoongAng Daily Newspaper. Her paper is titled, ‘A Hybrid Structure Dual-Path Step-Down Converter with 96.2% Peak Efficiency using 250mΩ Large-DCR Inductor’. Electronic devices require numerous chips and have a power converter to supply energy adequately. She proposed a new structure to enhance energy efficiency by combining inductors and capacitors. Enhancing energy efficiency can reduce energy loss, which prolongs battery hours and solves overheating of devices; for instance, energy loss leads to the overheating issue affecting phone chargers. This technology can be applied to various electronic devices, such as cell phones, laptops, and drones. Huh said, “Power has to go up in order to meet customers’ needs; however the overheating problem emerges during this process. This problem affects surrounding circuits and causes other issues, such as malfunctions of electronic devices. This technology may vary according to the conditions, but it can enhance energy efficiency up to 4%.”During the ceremony, about eight hundred million KRW worth cash prizes was conferred to 119 papers. KAIST (44 papers) and Gyeonggi Science High School (6 papers) received special awards given to the schools.
2018.02.12 View 11690 -
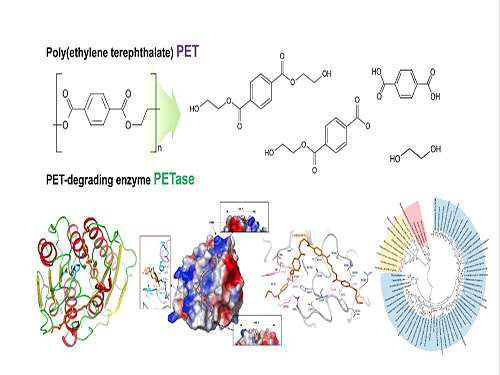 Structural Insight into the Molecular Mechanism of PET Degradation
A KAIST metabolic engineering research team has newly suggested a molecular mechanism showing superior degradability of poly ethylene terephthalate (PET).
This is the first report to simultaneously determine the 3D crystal structure of Ideonella sakaiensis PETase and develop the new variant with enhanced PET degradation.
Recently, diverse research projects are working to address the non-degradability of materials. A poly ethylene terephthalate (PET)-degrading bacterium called Ideonella sakaiensis was recently identified for the possible degradation and recycling of PET by Japanese team in Science journal (Yoshida et al., 2016). However, the detailed molecular mechanism of PET degradation has not been yet identified.
The team under Distinguished Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering and the team under Professor Kyung-Jin Kim of the Department of Biotechnology at Kyungpook National University conducted this research. The findings were published in Nature Communications on January 26.
This research predicts a special molecular mechanism based on the docking simulation between PETase and a PET alternative mimic substrate. Furthermore, they succeeded in constructing the variant for IsPETase with enhanced PET-degrading activity using structural-based protein engineering.
It is expected that the new approaches taken in this research can be background for further study of other enzymes capable of degrading not only PET but other plastics as well.
PET is very important source in our daily lives. However, PET after use causes tremendous contamination issues to our environment due to its non-biodegradability, which has been a major advantage of PET. Conventionally, PET is disposed of in landfills, using incineration, and sometimes recycling using chemical methods, which induces additional environmental pollution. Therefore, a new development for highly-efficient PET degrading enzymes is essential to degrade PET using bio-based eco-friendly methods.
Recently, a new bacterial species, Ideonella sakaiensis, which can use PET as a carbon source, was isolated. The PETase of I. sakaiensis (IsPETase) can degrade PET with relatively higher success than other PET-degrading enzymes. However, the detailed enzyme mechanism has not been elucidated, hindering further studies.
The research teams investigated how the substrate binds to the enzyme and which differences in enzyme structure result in significantly higher PET degrading activity compared with other cutinases and esterases, which make IsPETase highly attractive for industrial applications toward PET waste recycling.
Based on the 3D structure and related biochemical studies, they successfully predicted the reasons for extraordinary PET degrading activity of IsPETase and suggested other enzymes that can degrade PET with a newly-classified phylogenetic tree. The team proposed that 4 MHET moieties are the most properly matched substrates due to a cleft on structure even with the 10-20-mers for PET. This is meaningful in that it is the first docking simulation between PETase and PET, not its monomer.
Furthermore, they succeeded in developing a new variant with much higher PET-degrading activity using a crystal structure of this variant to show that the changed structure is better to accommodate PET substrates than wild type PETase, which will lead to developing further superior enzymes and constructing platforms for microbial plastic recycling.
Professor Lee said, “Environmental pollution from plastics remains one of the greatest challenges worldwide with the increasing consumption of plastics. We successfully constructed a new superior PET-degrading variant with the determination of a crystal structure of PETase and its degrading molecular mechanism. This novel technology will help further studies to engineer more superior enzymes with high efficiency in degrading. This will be the subject of our team’s ongoing research projects to address the global environmental pollution problem for next generation.”
This work was supported by the Technology Development Program to Solve Climate Changes on Systems Metabolic Engineering for Biorefineries (NRF-2012M1A2A2026556 and NRF-2012M1A2A2026557) from the Ministry of Science and ICT through the National Research Foundation of Korea. Further Contact: Dr. Sang Yup Lee, Distinguished Professor, KAIST, Daejeon, Korea (leesy@kaist.ac.kr, +82-42-350-3930)
(Figure: Structural insight into the molecular mechanism of poly(ethylene terephthalate) degradation and the phylogenetic tree of possible PET degrading enzymes. This schematic diagram shows the overall conceptualization for structural insight into the molecular mechanism of poly (ethylene terephthalate) degradation and the phylogenetic tree of possible PET degrading enzymes.)
2018.01.31 View 10335
Structural Insight into the Molecular Mechanism of PET Degradation
A KAIST metabolic engineering research team has newly suggested a molecular mechanism showing superior degradability of poly ethylene terephthalate (PET).
This is the first report to simultaneously determine the 3D crystal structure of Ideonella sakaiensis PETase and develop the new variant with enhanced PET degradation.
Recently, diverse research projects are working to address the non-degradability of materials. A poly ethylene terephthalate (PET)-degrading bacterium called Ideonella sakaiensis was recently identified for the possible degradation and recycling of PET by Japanese team in Science journal (Yoshida et al., 2016). However, the detailed molecular mechanism of PET degradation has not been yet identified.
The team under Distinguished Professor Sang Yup Lee of the Department of Chemical and Biomolecular Engineering and the team under Professor Kyung-Jin Kim of the Department of Biotechnology at Kyungpook National University conducted this research. The findings were published in Nature Communications on January 26.
This research predicts a special molecular mechanism based on the docking simulation between PETase and a PET alternative mimic substrate. Furthermore, they succeeded in constructing the variant for IsPETase with enhanced PET-degrading activity using structural-based protein engineering.
It is expected that the new approaches taken in this research can be background for further study of other enzymes capable of degrading not only PET but other plastics as well.
PET is very important source in our daily lives. However, PET after use causes tremendous contamination issues to our environment due to its non-biodegradability, which has been a major advantage of PET. Conventionally, PET is disposed of in landfills, using incineration, and sometimes recycling using chemical methods, which induces additional environmental pollution. Therefore, a new development for highly-efficient PET degrading enzymes is essential to degrade PET using bio-based eco-friendly methods.
Recently, a new bacterial species, Ideonella sakaiensis, which can use PET as a carbon source, was isolated. The PETase of I. sakaiensis (IsPETase) can degrade PET with relatively higher success than other PET-degrading enzymes. However, the detailed enzyme mechanism has not been elucidated, hindering further studies.
The research teams investigated how the substrate binds to the enzyme and which differences in enzyme structure result in significantly higher PET degrading activity compared with other cutinases and esterases, which make IsPETase highly attractive for industrial applications toward PET waste recycling.
Based on the 3D structure and related biochemical studies, they successfully predicted the reasons for extraordinary PET degrading activity of IsPETase and suggested other enzymes that can degrade PET with a newly-classified phylogenetic tree. The team proposed that 4 MHET moieties are the most properly matched substrates due to a cleft on structure even with the 10-20-mers for PET. This is meaningful in that it is the first docking simulation between PETase and PET, not its monomer.
Furthermore, they succeeded in developing a new variant with much higher PET-degrading activity using a crystal structure of this variant to show that the changed structure is better to accommodate PET substrates than wild type PETase, which will lead to developing further superior enzymes and constructing platforms for microbial plastic recycling.
Professor Lee said, “Environmental pollution from plastics remains one of the greatest challenges worldwide with the increasing consumption of plastics. We successfully constructed a new superior PET-degrading variant with the determination of a crystal structure of PETase and its degrading molecular mechanism. This novel technology will help further studies to engineer more superior enzymes with high efficiency in degrading. This will be the subject of our team’s ongoing research projects to address the global environmental pollution problem for next generation.”
This work was supported by the Technology Development Program to Solve Climate Changes on Systems Metabolic Engineering for Biorefineries (NRF-2012M1A2A2026556 and NRF-2012M1A2A2026557) from the Ministry of Science and ICT through the National Research Foundation of Korea. Further Contact: Dr. Sang Yup Lee, Distinguished Professor, KAIST, Daejeon, Korea (leesy@kaist.ac.kr, +82-42-350-3930)
(Figure: Structural insight into the molecular mechanism of poly(ethylene terephthalate) degradation and the phylogenetic tree of possible PET degrading enzymes. This schematic diagram shows the overall conceptualization for structural insight into the molecular mechanism of poly (ethylene terephthalate) degradation and the phylogenetic tree of possible PET degrading enzymes.)
2018.01.31 View 10335 -
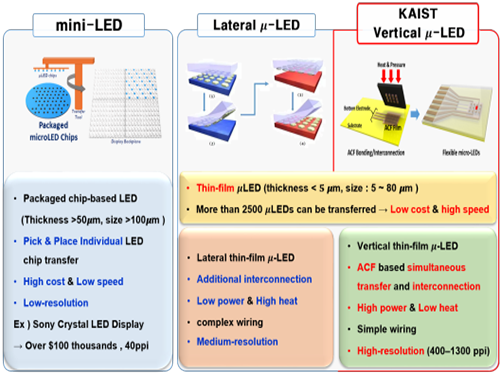 Developing Flexible Vertical Micro LED
A KAIST research team led by Professor Keon Jae Lee from the Department of Materials Science and Engineering and Professor Daesoo Kim from the Department of Biological Sciences has developed flexible vertical micro LEDs (f-VLEDs) using anisotropic conductive film (ACF)-based transfer and interconnection technology. The team also succeeded in controlling animal behavior via optogenetic stimulation of the f-VLEDs.
Flexible micro LEDs have become a strong candidate for the next-generation display due to their ultra-low power consumption, fast response speed, and excellent flexibility. However, the previous micro LED technology had critical issues such as poor device efficiency, low thermal reliability, and the lack of interconnection technology for high-resolution micro LED displays.
The research team has designed new transfer equipment and fabricated a f-VLED array (50ⅹ50) using simultaneous transfer and interconnection through the precise alignment of ACF bonding process. These f-VLEDs (thickness: 5 ㎛, size: below 80 ㎛) achieved optical power density (30 mW/mm2) three times higher than that of lateral micro LEDs, improving thermal reliability and lifetime by reducing heat generation within the thin film LEDs.
These f-VLEDs can be applied to optogenetics for controlling the behavior of neuron cells and brains. In contrast to the electrical stimulation that activates all of the neurons in brain, optogenetics can stimulate specific excitatory or inhibitory neurons within the localized cortical areas of the brain, which facilitates precise analysis, high-resolution mapping, and neuron modulation of animal brains. (Refer to the author’s previous ACS Nano paper of “Optogenetic Mapping of Functional Connectivity in Freely Moving Mice via Insertable Wrapping Electrode Array Beneath the Skull.” )
In this work, they inserted the innovative f-VLEDs into the narrow space between the skull and the brain surface and succeeded in controlling mouse behavior by illuminating motor neurons on two-dimensional cortical areas located deep below the brain surface.
Professor Lee said, “The flexible vertical micro LED can be used in low-power smart watches, mobile displays, and wearable lighting. In addition, these flexible optoelectronic devices are suitable for biomedical applications such as brain science, phototherapeutic treatment, and contact lens biosensors.”
He recently established a startup company ( FRONICS Inc. ) based on micro LED technology and is looking for global partnerships for commercialization. This result entitled “ Optogenetic Control of Body Movements via Flexible Vertical Light-Emitting Diodes on Brain Surface ” was published in the February 2018 issue of Nano Energy.
Figure 1. Comparison of μ-LEDs Technology
2018.01.29 View 14953
Developing Flexible Vertical Micro LED
A KAIST research team led by Professor Keon Jae Lee from the Department of Materials Science and Engineering and Professor Daesoo Kim from the Department of Biological Sciences has developed flexible vertical micro LEDs (f-VLEDs) using anisotropic conductive film (ACF)-based transfer and interconnection technology. The team also succeeded in controlling animal behavior via optogenetic stimulation of the f-VLEDs.
Flexible micro LEDs have become a strong candidate for the next-generation display due to their ultra-low power consumption, fast response speed, and excellent flexibility. However, the previous micro LED technology had critical issues such as poor device efficiency, low thermal reliability, and the lack of interconnection technology for high-resolution micro LED displays.
The research team has designed new transfer equipment and fabricated a f-VLED array (50ⅹ50) using simultaneous transfer and interconnection through the precise alignment of ACF bonding process. These f-VLEDs (thickness: 5 ㎛, size: below 80 ㎛) achieved optical power density (30 mW/mm2) three times higher than that of lateral micro LEDs, improving thermal reliability and lifetime by reducing heat generation within the thin film LEDs.
These f-VLEDs can be applied to optogenetics for controlling the behavior of neuron cells and brains. In contrast to the electrical stimulation that activates all of the neurons in brain, optogenetics can stimulate specific excitatory or inhibitory neurons within the localized cortical areas of the brain, which facilitates precise analysis, high-resolution mapping, and neuron modulation of animal brains. (Refer to the author’s previous ACS Nano paper of “Optogenetic Mapping of Functional Connectivity in Freely Moving Mice via Insertable Wrapping Electrode Array Beneath the Skull.” )
In this work, they inserted the innovative f-VLEDs into the narrow space between the skull and the brain surface and succeeded in controlling mouse behavior by illuminating motor neurons on two-dimensional cortical areas located deep below the brain surface.
Professor Lee said, “The flexible vertical micro LED can be used in low-power smart watches, mobile displays, and wearable lighting. In addition, these flexible optoelectronic devices are suitable for biomedical applications such as brain science, phototherapeutic treatment, and contact lens biosensors.”
He recently established a startup company ( FRONICS Inc. ) based on micro LED technology and is looking for global partnerships for commercialization. This result entitled “ Optogenetic Control of Body Movements via Flexible Vertical Light-Emitting Diodes on Brain Surface ” was published in the February 2018 issue of Nano Energy.
Figure 1. Comparison of μ-LEDs Technology
2018.01.29 View 14953 -
 Three Professors Named KAST Fellows
(Professor Dan Keun Sung at the center)
(Professor Y.H. Cho at the center)
(Professor K.H. Cho at the center)
The Korean Academy of Science and Technology (KAST) inducted three KAIST professors as fellows at the New Year’s ceremony held at KAST on January 12. They were among the 24 newly elected fellows of the most distinguished academy in Korea. The new fellows are Professor Dan Keun Sung of the School of Electrical Engineering, Professor Kwang-Hyun Cho of the Department of Bio and Brain Engineering, and Professor Yong-Hoon Cho of the Department of Physics.
Professor Sung was recognized for his lifetime academic achievements in fields related with network protocols and energy ICT. He also played a crucial role in launching the Korean satellites KITSAT-1,2,3 and the establishment of the Satellite Technology Research Center at KAIST.
Professor Y.H.Cho has been a pioneer in the field of low-dimensional semiconductor-powered quantum photonics that enables quantum optical research in solid state. He has been recognized as a renowned scholar in this field internationally.
Professor K.H.Cho has conducted original research that combines IT and BT in systems biology and has applied novel technologies of electronic modeling and computer simulation analysis for investigating complex life sciences. Professor Cho, who is in his 40s, is the youngest fellow among the newly inducted fellows.
2018.01.16 View 17608
Three Professors Named KAST Fellows
(Professor Dan Keun Sung at the center)
(Professor Y.H. Cho at the center)
(Professor K.H. Cho at the center)
The Korean Academy of Science and Technology (KAST) inducted three KAIST professors as fellows at the New Year’s ceremony held at KAST on January 12. They were among the 24 newly elected fellows of the most distinguished academy in Korea. The new fellows are Professor Dan Keun Sung of the School of Electrical Engineering, Professor Kwang-Hyun Cho of the Department of Bio and Brain Engineering, and Professor Yong-Hoon Cho of the Department of Physics.
Professor Sung was recognized for his lifetime academic achievements in fields related with network protocols and energy ICT. He also played a crucial role in launching the Korean satellites KITSAT-1,2,3 and the establishment of the Satellite Technology Research Center at KAIST.
Professor Y.H.Cho has been a pioneer in the field of low-dimensional semiconductor-powered quantum photonics that enables quantum optical research in solid state. He has been recognized as a renowned scholar in this field internationally.
Professor K.H.Cho has conducted original research that combines IT and BT in systems biology and has applied novel technologies of electronic modeling and computer simulation analysis for investigating complex life sciences. Professor Cho, who is in his 40s, is the youngest fellow among the newly inducted fellows.
2018.01.16 View 17608