research
- A computer simulation program “iBridge” was developed at KAIST that can put together microbial cell factories quickly and efficiently to produce cosmetics and food additives, and raw materials for nylons
- Eco-friendly and sustainable fermentation process to establish an alternative to chemical plants
As climate change and environmental concerns intensify, sustainable microbial cell factories garner significant attention as candidates to replace chemical plants. To develop microorganisms to be used in the microbial cell factories, it is crucial to modify their metabolic processes to induce efficient target chemical production by modulating its gene expressions. Yet, the challenge persists in determining which gene expressions to amplify and suppress, and the experimental verification of these modification targets is a time- and resource-intensive process even for experts. The challenges were addressed by a team of researchers at KAIST (President Kwang-Hyung Lee) led by Distinguished Professor Sang Yup Lee.
It was announced on the 9th by the school that a method for building a microbial factory at low cost, quickly and efficiently, was presented by a novel computer simulation program developed by the team under Professor Lee’s guidance, which is named “iBridge”. This innovative system is designed to predict gene targets to either overexpress or downregulate in the goal of producing a desired compound to enable the cost-effective and efficient construction of microbial cell factories specifically tailored for producing the chemical compound in demand from renewable biomass.
Systems metabolic engineering is a field of research and engineering pioneered by KAIST’s Distinguished Professor Sang Yup Lee that seeks to produce valuable compounds in industrial demands using microorganisms that are re-configured by a combination of methods including, but not limited to, metabolic engineering, synthetic biology, systems biology, and fermentation engineering.
In order to improve microorganisms’ capability to produce useful compounds, it is essential to delete, suppress, or overexpress microbial genes. However, it is difficult even for the experts to identify the gene targets to modify without experimental confirmations for each of them, which can take up immeasurable amount of time and resources.
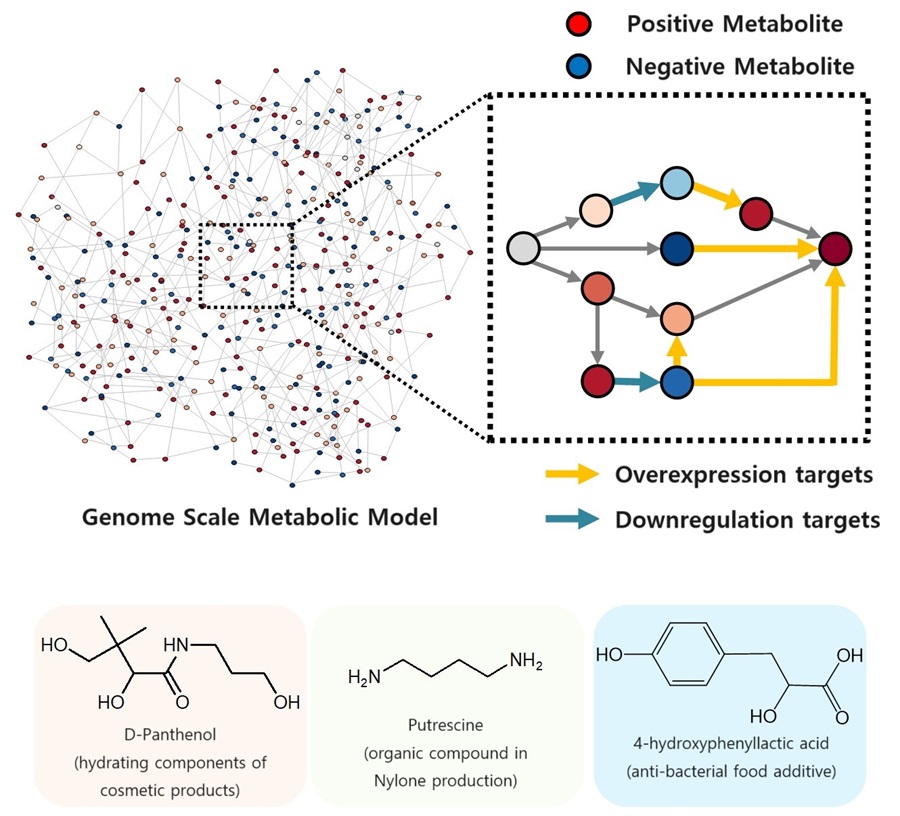
The newly developed iBridge identifies positive and negative metabolites within cells, which exert positive and/or negative impact on formation of the products, by calculating the sum of covariances of their outgoing (consuming) reaction fluxes for a target chemical. Subsequently, it pinpoints "bridge" reactions responsible for converting negative metabolites into positive ones as candidates for overexpression, while identifying the opposites as targets for downregulation.
The research team successfully utilized the iBridge simulation to establish E. coli microbial cell factories each capable of producing three of the compounds that are in high demands at a production capacity that has not been reported around the world. They developed E. coli strains that can each produce panthenol, a moisturizing agent found in many cosmetics, putrescine, which is one of the key components in nylon production, and 4-hydroxyphenyllactic acid, an anti-bacterial food additive. In addition to these three compounds, the study presents predictions for overexpression and suppression genes to construct microbial factories for 298 other industrially valuable compounds.
Dr. Youngjoon Lee, the co-first author of this paper from KAIST, emphasized the accelerated construction of various microbial factories the newly developed simulation enabled. He stated, "With the use of this simulation, multiple microbial cell factories have been established significantly faster than it would have been using the conventional methods. Microbial cell factories producing a wider range of valuable compounds can now be constructed quickly using this technology."
Professor Sang Yup Lee said, "Systems metabolic engineering is a crucial technology for addressing the current climate change issues." He added, "This simulation could significantly expedite the transition from resorting to conventional chemical factories to utilizing environmentally friendly microbial factories."
< Figure. Conceptual diagram of the flow of iBridge simulation >
The team’s work on iBridge is described in a paper titled "Genome-Wide Identification of Overexpression and Downregulation Gene Targets Based on the Sum of Covariances of the Outgoing Reaction Fluxes" written by Dr. Won Jun Kim, and Dr. Youngjoon Lee of the Bioprocess Research Center and Professors Hyun Uk Kim and Sang Yup Lee of the Department of Chemical and Biomolecular Engineering of KAIST. The paper was published via peer-review on the 6th of November on “Cell Systems” by Cell Press.
This research was conducted with the support from the Development of Platform Technologies of Microbial Cell Factories for the Next-generation Biorefineries Project (Project Leader: Distinguished Professor Sang Yup Lee, KAIST) and Development of Platform Technology for the Production of Novel Aromatic Bioplastic using Microbial Cell Factories Project (Project Leader: Research Professor So Young Choi, KAIST) of the Korean Ministry of Science and ICT.
-
people VP Sang Yup Lee Honored with the Pony Chung Innovation Award
Vice President for Research Sang Yup Lee became the recipient of the Innovation Award by the Pony Chung Foundation that was established to honor the late Se-yung Chung, the former chairman of Hyundai Development Company. He will receive 200 million KRW in prize money. Chairman Chung developed Korea’s first domestically manufactured automobile, ‘Pony,’ in the mid-1970s that became the cornerstone of Korea’s auto industry today. Distinguished Professor Lee, from the Depa
2021-07-13 -
research Deep Learning-Powered 'DeepEC' Helps Accurately Understand Enzyme Functions
(Figure: Overall scheme of DeepEC) A deep learning-powered computational framework, ‘DeepEC,’ will allow the high-quality and high-throughput prediction of enzyme commission numbers, which is essential for the accurate understanding of enzyme functions. A team of Dr. Jae Yong Ryu, Professor Hyun Uk Kim, and Distinguished Professor Sang Yup Lee at KAIST reported the computational framework powered by deep learning that predicts enzyme commission (EC) numbers with high precisio
2019-07-09 -
research Efficiently Producing Fatty Acids and Biofuels from Glucose
Researchers have presented a new strategy for efficiently producing fatty acids and biofuels that can transform glucose and oleaginous microorganisms into microbial diesel fuel, with one-step direct fermentative production. The newly developed strain, created by Distinguished Professor Sang Yup Lee and his team, showed the highest efficiency in producing fatty acids and biodiesels ever reported. It will be expected to serve as a new platform to sustainably produce a wide array of fatty acid
2019-06-19 -
research Engineered Microbial Production of Grape Flavoring
(Image 1: Engineered bacteria that produce grape flavoring.) Researchers report a microbial method for producing an artificial grape flavor. Methyl anthranilate (MANT) is a common grape flavoring and odorant compound currently produced through a petroleum-based process that uses large volumes of toxic acid catalysts. Professor Sang-Yup Lee’s team at the Department of Chemical and Biomolecular Engineering demonstrated production of MANT, a naturally occurring compound, via engin
2019-05-15 -
people Top 10 Emerging Technologies by World Economic Forum
The World Economic Forum’s Meta-Council on Emerging Technologies announced its annual list of breakthrough technologies, the “Top 10 Emerging Technologies of 2016,” on June 23, 2016. The Meta-Council chose the top ten technologies based on the technologies’ potential to improve lives, transform industries, and safeguard the planet. The research field of systems metabolic engineering, founded by Distinguished Professor Sang Yup Lee of the Chemical and Biomolecular Engineer
2016-06-27