carcinogenic
-
 Hidden Mechanism for the Suppression of Colon Cancer Identified
Published in Cell Reports : cells at the risk of causing colorectal cancer due to genetic mutation are discharged outside the colon tissue
Korean researchers have successfully identified the cancer inhibitory mechanism of the colon tissue. The discovery of the inherent defense mechanism of the colon tissues is expected to provide understanding of the cause of colorectal cancer.
The research was led by Kwang-Hyun Cho, a professor of Bio and Brain Engineering at KAIST (corresponding author) and participated by Dr. Jehun Song (the first author), as well as Dr. Owen Sansom, David Huels, and Rachel Ridgway from the Beatson Institute for Cancer Research in the UK and Dr. Walter Kolch from Conway Institute in Ireland.
The research was funded by the Ministry of Science, ICT and Future Planning and the National Research Foundation of Korea, and its results were published in the 28th March online edition of Cell Reports under the title of “The APC network regulates the removal of mutated cells from colonic crypts.”
The organism can repair damaged tissues by itself, but genetic mutations, which may cause cancer, can occur in the process of cell division s for the repair. The rapid cell division s and toxic substances from the digestive process cause a problem especially in colon crypt that has a high probability for genetic mutation.
The research team was able to find out that the colon tissues prevent cancer by rapidly discharging carcinogenic cells with genetic mutations from the colon crypt durin ga frequent tissue repair process.
This defense mechanism, which inhibits abnormal cell division s by reducing the time mutated cells reside in the crypt, is inherent in the colon.
Extensive mathematical simulation results show that the mutated cells with enhanced Wnt signaling acquire increased adhesion in comparison to the normal cells, which therefore move rapidly toward the upper part of the crypt and are discharged more easily.
If beta-catenine, the key factor in Wnt signal transduction pathway, is not degraded due to genetic mutation, the accumulated beta-catenine activates cell proliferation and increases cell adhesion. The special environment of crypt tissue and the tendency of the cells with similar adhesion to aggregate will therefore discharge the mutated cell, hence maintaining the tissue homeostasis.
In vivo experiment with a mouse model confirms the simulation results that, in the case of abnormal crypt, the cells with high proliferation in fact move slower.
Professor Cho said, “This research has identified that multicellular organism is exquisitely designed to maintain the tissue homeostasis despite abnormal cell mutation. This also proves the systems biology research, which is a convergence of information technology and bio-technology , can discover hidden mechanisms behind complex biological phenomena.”
Crypt: Epithelium, consisting of approximately 2,000 cells, forms a colon surface in the shape of a cave.
Wnt Signaling: A signal transduction pathway involved in the proliferation and differentiation of cells that are particularly important for the embryonic development and management of adult tissue homeostasis.
2014.04.17 View 12450
Hidden Mechanism for the Suppression of Colon Cancer Identified
Published in Cell Reports : cells at the risk of causing colorectal cancer due to genetic mutation are discharged outside the colon tissue
Korean researchers have successfully identified the cancer inhibitory mechanism of the colon tissue. The discovery of the inherent defense mechanism of the colon tissues is expected to provide understanding of the cause of colorectal cancer.
The research was led by Kwang-Hyun Cho, a professor of Bio and Brain Engineering at KAIST (corresponding author) and participated by Dr. Jehun Song (the first author), as well as Dr. Owen Sansom, David Huels, and Rachel Ridgway from the Beatson Institute for Cancer Research in the UK and Dr. Walter Kolch from Conway Institute in Ireland.
The research was funded by the Ministry of Science, ICT and Future Planning and the National Research Foundation of Korea, and its results were published in the 28th March online edition of Cell Reports under the title of “The APC network regulates the removal of mutated cells from colonic crypts.”
The organism can repair damaged tissues by itself, but genetic mutations, which may cause cancer, can occur in the process of cell division s for the repair. The rapid cell division s and toxic substances from the digestive process cause a problem especially in colon crypt that has a high probability for genetic mutation.
The research team was able to find out that the colon tissues prevent cancer by rapidly discharging carcinogenic cells with genetic mutations from the colon crypt durin ga frequent tissue repair process.
This defense mechanism, which inhibits abnormal cell division s by reducing the time mutated cells reside in the crypt, is inherent in the colon.
Extensive mathematical simulation results show that the mutated cells with enhanced Wnt signaling acquire increased adhesion in comparison to the normal cells, which therefore move rapidly toward the upper part of the crypt and are discharged more easily.
If beta-catenine, the key factor in Wnt signal transduction pathway, is not degraded due to genetic mutation, the accumulated beta-catenine activates cell proliferation and increases cell adhesion. The special environment of crypt tissue and the tendency of the cells with similar adhesion to aggregate will therefore discharge the mutated cell, hence maintaining the tissue homeostasis.
In vivo experiment with a mouse model confirms the simulation results that, in the case of abnormal crypt, the cells with high proliferation in fact move slower.
Professor Cho said, “This research has identified that multicellular organism is exquisitely designed to maintain the tissue homeostasis despite abnormal cell mutation. This also proves the systems biology research, which is a convergence of information technology and bio-technology , can discover hidden mechanisms behind complex biological phenomena.”
Crypt: Epithelium, consisting of approximately 2,000 cells, forms a colon surface in the shape of a cave.
Wnt Signaling: A signal transduction pathway involved in the proliferation and differentiation of cells that are particularly important for the embryonic development and management of adult tissue homeostasis.
2014.04.17 View 12450 -
 The new era of personalized cancer diagnosis and treatment
Professor Tae-Young Yoon
- Succeeded in observing carcinogenic protein at the molecular level
- “Paved the way to customized cancer treatment through accurate analysis of carcinogenic protein”
The joint KAIST research team of Professor Tae Young Yoon of the Department of Physics and Professor Won Do Huh of the Department of Biological Sciences have developed the technology to monitor characteristics of carcinogenic protein in cancer tissue – for the first time in the world.
The technology makes it possible to analyse the mechanism of cancer development through a small amount of carcinogenic protein from a cancer patient. Therefore, a personalised approach to diagnosis and treatment using the knowledge of the specific mechanism of cancer development in the patient may be possible in the future.
Until recently, modern medicine could only speculate on the cause of cancer through statistics. Although developed countries, such as the United States, are known to use a large sequencing technology that analyses the patient’s DNA, identification of the interactions between proteins responsible for causing cancer remained an unanswered question for a long time in medicine.
Firstly, Professor Yoon’s research team has developed a fluorescent microscope that can observe even a single molecule. Then, the “Immunoprecipitation method”, a technology to extract a specific protein exploiting the high affinity between antigens and antibodies was developed. Using this technology and the microscope, “Real-Time Single Molecule co-Immunoprecipitation Method” was created. In this way, the team succeeded in observing the interactions between carcinogenic and other proteins at a molecular level, in real time.
To validate the developed technology, the team investigated Ras, a carcinogenic protein; its mutation statistically is known to cause around 30% of cancers.
The experimental results confirmed that 30-50% of Ras protein was expressed in mouse tumour and human cancer cells. In normal cells, less than 5% of Ras protein was expressed. Thus, the experiment showed that unusual increase in activation of Ras protein induces cancer.
The increase in the ratio of active Ras protein can be inferred from existing research data but the measurement of specific numerical data has never been done before.
The team suggested a new molecular level diagnosis technique of identifying the progress of cancer in patients through measuring the percentage of activated carcinogenic protein in cancer tissue.
Professor Yoon Tae-young said, “This newly developed technology does not require a separate procedure of protein expression or refining, hence the existing proteins in real biological tissues or cancer cells can be observed directly.” He also said, “Since carcinogenic protein can be analyzed accurately, it has opened up the path to customized cancer treatment in the future.”
“Since the observation is possible on a molecular level, the technology confers the advantage that researchers can carry out various examinations on a small sample of the cancer patient.” He added, “The clinical trial will start in December 2012 and in a few years customized cancer diagnosis and treatment will be possible.”
Meanwhile, the research has been published in Nature Communications (February 19). Many researchers from various fields have participated, regardless of the differences in their speciality, and successfully produced interdisciplinary research. Professor Tae Young Yoon of the Department of Physics and Professors Dae Sik Lim and Won Do Huh of Biological Sciences at KAIST, and Professor Chang Bong Hyun of Computational Science of KIAS contributed to developing the technique.
Figure 1: Schematic diagram of observed interactions at the molecular level in real time using fluorescent microscope. The carcinogenic protein from a mouse tumour is fixed on the microchip, and its molecular characteristics are observed live.
Figure 2: Molecular interaction data using a molecular level fluorescent microscope. A signal in the form of spike is shown when two proteins combine. This is monitored live using an Electron Multiplying Charge Coupled Device (EMCCD). It shows signal results in bright dots.
An organism has an immune system as a defence mechanism to foreign intruders. The immune system is activated when unwanted pathogens or foreign protein are in the body. Antibodies form in recognition of the specific antigen to protect itself. Organisms evolved to form antibodies with high specificity to a certain antigen. Antibodies only react to its complementary antigens. The field of molecular biology uses the affinity between antigens and antibodies to extract specific proteins; a technology called immunoprecipitation. Even in a mixture of many proteins, the protein sought can be extracted using antibodies. Thus immunoprecipitation is widely used to detect pathogens or to extract specific proteins.
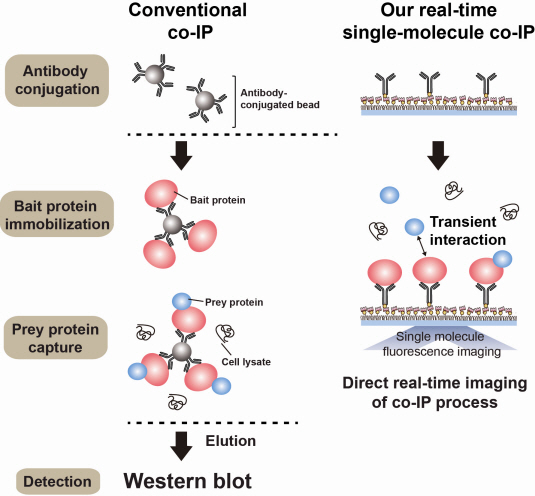
Technology co-IP is a well-known example that uses immunoprecipitation. The research on interactions between proteins uses co-IP in general. The basis of fixing the antigen on the antibody to extract antigen protein is the same as immunoprecipitation. Then, researchers inject and observe its reaction with the partner protein to observe the interactions and precipitate the antibodies. If the reaction occurs, the partner protein will be found with the antibodies in the precipitations. If not, then the partner protein will not be found. This shows that the two proteins interact.
However, the traditional co-IP can be used to infer the interactions between the two proteins although the information of the dynamics on how the reaction occurs is lost. To overcome these shortcomings, the Real-Time Single Molecule co-IP Method enables observation on individual protein level in real time. Therefore, the significance of the new technique is in making observation of interactions more direct and quantitative.
Additional Figure 1: Comparison between Conventional co-IP and Real-Time Single Molecule co-IP
2013.04.01 View 19433
The new era of personalized cancer diagnosis and treatment
Professor Tae-Young Yoon
- Succeeded in observing carcinogenic protein at the molecular level
- “Paved the way to customized cancer treatment through accurate analysis of carcinogenic protein”
The joint KAIST research team of Professor Tae Young Yoon of the Department of Physics and Professor Won Do Huh of the Department of Biological Sciences have developed the technology to monitor characteristics of carcinogenic protein in cancer tissue – for the first time in the world.
The technology makes it possible to analyse the mechanism of cancer development through a small amount of carcinogenic protein from a cancer patient. Therefore, a personalised approach to diagnosis and treatment using the knowledge of the specific mechanism of cancer development in the patient may be possible in the future.
Until recently, modern medicine could only speculate on the cause of cancer through statistics. Although developed countries, such as the United States, are known to use a large sequencing technology that analyses the patient’s DNA, identification of the interactions between proteins responsible for causing cancer remained an unanswered question for a long time in medicine.
Firstly, Professor Yoon’s research team has developed a fluorescent microscope that can observe even a single molecule. Then, the “Immunoprecipitation method”, a technology to extract a specific protein exploiting the high affinity between antigens and antibodies was developed. Using this technology and the microscope, “Real-Time Single Molecule co-Immunoprecipitation Method” was created. In this way, the team succeeded in observing the interactions between carcinogenic and other proteins at a molecular level, in real time.
To validate the developed technology, the team investigated Ras, a carcinogenic protein; its mutation statistically is known to cause around 30% of cancers.
The experimental results confirmed that 30-50% of Ras protein was expressed in mouse tumour and human cancer cells. In normal cells, less than 5% of Ras protein was expressed. Thus, the experiment showed that unusual increase in activation of Ras protein induces cancer.
The increase in the ratio of active Ras protein can be inferred from existing research data but the measurement of specific numerical data has never been done before.
The team suggested a new molecular level diagnosis technique of identifying the progress of cancer in patients through measuring the percentage of activated carcinogenic protein in cancer tissue.
Professor Yoon Tae-young said, “This newly developed technology does not require a separate procedure of protein expression or refining, hence the existing proteins in real biological tissues or cancer cells can be observed directly.” He also said, “Since carcinogenic protein can be analyzed accurately, it has opened up the path to customized cancer treatment in the future.”
“Since the observation is possible on a molecular level, the technology confers the advantage that researchers can carry out various examinations on a small sample of the cancer patient.” He added, “The clinical trial will start in December 2012 and in a few years customized cancer diagnosis and treatment will be possible.”
Meanwhile, the research has been published in Nature Communications (February 19). Many researchers from various fields have participated, regardless of the differences in their speciality, and successfully produced interdisciplinary research. Professor Tae Young Yoon of the Department of Physics and Professors Dae Sik Lim and Won Do Huh of Biological Sciences at KAIST, and Professor Chang Bong Hyun of Computational Science of KIAS contributed to developing the technique.
Figure 1: Schematic diagram of observed interactions at the molecular level in real time using fluorescent microscope. The carcinogenic protein from a mouse tumour is fixed on the microchip, and its molecular characteristics are observed live.
Figure 2: Molecular interaction data using a molecular level fluorescent microscope. A signal in the form of spike is shown when two proteins combine. This is monitored live using an Electron Multiplying Charge Coupled Device (EMCCD). It shows signal results in bright dots.
An organism has an immune system as a defence mechanism to foreign intruders. The immune system is activated when unwanted pathogens or foreign protein are in the body. Antibodies form in recognition of the specific antigen to protect itself. Organisms evolved to form antibodies with high specificity to a certain antigen. Antibodies only react to its complementary antigens. The field of molecular biology uses the affinity between antigens and antibodies to extract specific proteins; a technology called immunoprecipitation. Even in a mixture of many proteins, the protein sought can be extracted using antibodies. Thus immunoprecipitation is widely used to detect pathogens or to extract specific proteins.
Technology co-IP is a well-known example that uses immunoprecipitation. The research on interactions between proteins uses co-IP in general. The basis of fixing the antigen on the antibody to extract antigen protein is the same as immunoprecipitation. Then, researchers inject and observe its reaction with the partner protein to observe the interactions and precipitate the antibodies. If the reaction occurs, the partner protein will be found with the antibodies in the precipitations. If not, then the partner protein will not be found. This shows that the two proteins interact.
However, the traditional co-IP can be used to infer the interactions between the two proteins although the information of the dynamics on how the reaction occurs is lost. To overcome these shortcomings, the Real-Time Single Molecule co-IP Method enables observation on individual protein level in real time. Therefore, the significance of the new technique is in making observation of interactions more direct and quantitative.
Additional Figure 1: Comparison between Conventional co-IP and Real-Time Single Molecule co-IP
2013.04.01 View 19433