AND
-
 KAIST Develops AI That Automatically Designs Optimal Drug Candidates for Cancer-Targeting Mutations
< (From left) Ph.D candidate Wonho Zhung, Ph.D cadidate Joongwon Lee , Prof. Woo Young Kim , Ph.D candidate Jisu Seo >
Traditional drug development methods involve identifying a target protin (e.g., a cancer cell receptor) that causes disease, and then searching through countless molecular candidates (potential drugs) that could bind to that protein and block its function. This process is costly, time-consuming, and has a low success rate. KAIST researchers have developed an AI model that, using only information about the target protein, can design optimal drug candidates without any prior molecular data—opening up new possibilities for drug discovery.
KAIST (President Kwang Hyung Lee) announced on the 10th that a research team led by Professor Woo Youn Kim in the Department of Chemistry has developed an AI model named BInD (Bond and Interaction-generating Diffusion model), which can design and optimize drug candidate molecules tailored to a protein’s structure alone—without needing prior information about binding molecules. The model also predicts the binding mechanism (non-covalent interactions) between the drug and the target protein.
The core innovation of this technology lies in its “simultaneous design” approach. Previous AI models either focused on generating molecules or separately evaluating whether the generated molecule could bind to the target protein. In contrast, this new model considers the binding mechanism between the molecule and the protein during the generation process, enabling comprehensive design in one step. Since it pre-accounts for critical factors in protein-ligand binding, it has a much higher likelihood of generating effective and stable molecules. The generation process visually demonstrates how types and positions of atoms, covalent bonds, and interactions are created simultaneously to fit the protein’s binding site.
<Figure 1. Schematic of the diffusion model developed by the research team, which generates molecular structures and non-covalent interactions based on protein structures. Starting from a noise distribution, the model gradually removes noise (via reverse diffusion) to restore the atom positions, types, covalent bond types, and interaction types, thereby generating molecules. Interacting patterns are extracted from prior knowledge of known binding molecules or proteins, and through an inpainting technique, these patterns are kept fixed during the reverse diffusion process to guide the molecular generation.>
Moreover, this model is designed to meet multiple essential drug design criteria simultaneously—such as target binding affinity, drug-like properties, and structural stability. Traditional models often optimized for only one or two goals at the expense of others, but this new model balances various objectives, significantly enhancing its practical applicability.
The research team explained that the AI operates based on a “diffusion model”—a generative approach where a structure becomes increasingly refined from a random state. This is the same type of model used in AlphaFold 3, the 2024 Nobel Chemistry Prize-winning tool for protein-ligand structure generation, which has already demonstrated high efficiency.
Unlike AlphaFold 3, which provides spatial coordinates for atom positions, this study introduced a knowledge-based guide grounded in actual chemical laws—such as bond lengths and protein-ligand distances—enabling more chemically realistic structure generation.
<Figure 2. (Left) Target protein and the original bound molecule; (Right) Examples of molecules designed using the model developed in this study. The values for protein binding affinity (Vina), drug-likeness (QED), and synthetic accessibility (SA) are shown at the bottom.>
Additionally, the team applied an optimization strategy where outstanding binding patterns from prior results are reused. This allowed the model to generate even better drug candidates without additional training. Notably, the AI successfully produced molecules that selectively bind to the mutated residues of EGFR, a cancer-related target protein.
This study is also meaningful because it advances beyond the team’s previous research, which required prior input about the molecular conditions for the interaction pattern of protein binding.
Professor Woo Youn Kim commented that “the newly developed AI can learn and understand the key features required for strong binding to a target protein, and design optimal drug candidate molecules—even without any prior input. This could significantly shift the paradigm of drug development.” He added, “Since this technology generates molecular structures based on principles of chemical interactions, it is expected to enable faster and more reliable drug development.”
Joongwon Lee and Wonho Zhung, PhD students in the Department of Chemistry, participated as co-first authors of this study. The research results were published in the international journal Advanced Science (IF = 14.1) on July 11.
● Paper Title: BInD: Bond and Interaction-Generating Diffusion Model for Multi-Objective Structure-Based Drug Design
● DOI: 10.1002/advs.202502702
This research was supported by the National Research Foundation of Korea and the Ministry of Health and Welfare.
2025.08.12 View 22
KAIST Develops AI That Automatically Designs Optimal Drug Candidates for Cancer-Targeting Mutations
< (From left) Ph.D candidate Wonho Zhung, Ph.D cadidate Joongwon Lee , Prof. Woo Young Kim , Ph.D candidate Jisu Seo >
Traditional drug development methods involve identifying a target protin (e.g., a cancer cell receptor) that causes disease, and then searching through countless molecular candidates (potential drugs) that could bind to that protein and block its function. This process is costly, time-consuming, and has a low success rate. KAIST researchers have developed an AI model that, using only information about the target protein, can design optimal drug candidates without any prior molecular data—opening up new possibilities for drug discovery.
KAIST (President Kwang Hyung Lee) announced on the 10th that a research team led by Professor Woo Youn Kim in the Department of Chemistry has developed an AI model named BInD (Bond and Interaction-generating Diffusion model), which can design and optimize drug candidate molecules tailored to a protein’s structure alone—without needing prior information about binding molecules. The model also predicts the binding mechanism (non-covalent interactions) between the drug and the target protein.
The core innovation of this technology lies in its “simultaneous design” approach. Previous AI models either focused on generating molecules or separately evaluating whether the generated molecule could bind to the target protein. In contrast, this new model considers the binding mechanism between the molecule and the protein during the generation process, enabling comprehensive design in one step. Since it pre-accounts for critical factors in protein-ligand binding, it has a much higher likelihood of generating effective and stable molecules. The generation process visually demonstrates how types and positions of atoms, covalent bonds, and interactions are created simultaneously to fit the protein’s binding site.
<Figure 1. Schematic of the diffusion model developed by the research team, which generates molecular structures and non-covalent interactions based on protein structures. Starting from a noise distribution, the model gradually removes noise (via reverse diffusion) to restore the atom positions, types, covalent bond types, and interaction types, thereby generating molecules. Interacting patterns are extracted from prior knowledge of known binding molecules or proteins, and through an inpainting technique, these patterns are kept fixed during the reverse diffusion process to guide the molecular generation.>
Moreover, this model is designed to meet multiple essential drug design criteria simultaneously—such as target binding affinity, drug-like properties, and structural stability. Traditional models often optimized for only one or two goals at the expense of others, but this new model balances various objectives, significantly enhancing its practical applicability.
The research team explained that the AI operates based on a “diffusion model”—a generative approach where a structure becomes increasingly refined from a random state. This is the same type of model used in AlphaFold 3, the 2024 Nobel Chemistry Prize-winning tool for protein-ligand structure generation, which has already demonstrated high efficiency.
Unlike AlphaFold 3, which provides spatial coordinates for atom positions, this study introduced a knowledge-based guide grounded in actual chemical laws—such as bond lengths and protein-ligand distances—enabling more chemically realistic structure generation.
<Figure 2. (Left) Target protein and the original bound molecule; (Right) Examples of molecules designed using the model developed in this study. The values for protein binding affinity (Vina), drug-likeness (QED), and synthetic accessibility (SA) are shown at the bottom.>
Additionally, the team applied an optimization strategy where outstanding binding patterns from prior results are reused. This allowed the model to generate even better drug candidates without additional training. Notably, the AI successfully produced molecules that selectively bind to the mutated residues of EGFR, a cancer-related target protein.
This study is also meaningful because it advances beyond the team’s previous research, which required prior input about the molecular conditions for the interaction pattern of protein binding.
Professor Woo Youn Kim commented that “the newly developed AI can learn and understand the key features required for strong binding to a target protein, and design optimal drug candidate molecules—even without any prior input. This could significantly shift the paradigm of drug development.” He added, “Since this technology generates molecular structures based on principles of chemical interactions, it is expected to enable faster and more reliable drug development.”
Joongwon Lee and Wonho Zhung, PhD students in the Department of Chemistry, participated as co-first authors of this study. The research results were published in the international journal Advanced Science (IF = 14.1) on July 11.
● Paper Title: BInD: Bond and Interaction-Generating Diffusion Model for Multi-Objective Structure-Based Drug Design
● DOI: 10.1002/advs.202502702
This research was supported by the National Research Foundation of Korea and the Ministry of Health and Welfare.
2025.08.12 View 22 -
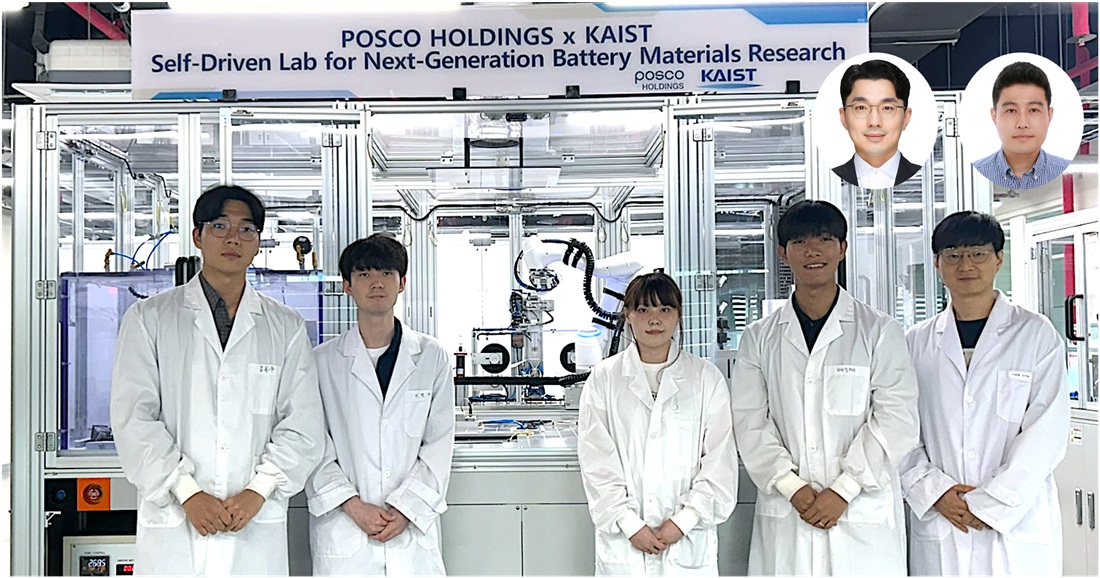 Material Innovation Realized with Robotic Arms and AI, Without Human Researchers
<(From Left) M.S candidate Dongwoo Kim from KAIST, Ph.D candidate Hyun-Gi Lee from KAIST, Intern Yeham Kang from KAIST, M.S candidate Seongjae Bae from KAIST, Professor Dong-Hwa Seo from KAIST, (From top right, from left) Senior Researcher Inchul Park from POSCO Holdings, Senior Researcher Jung Woo Park, senior researcher from POSCO Holdings>
A joint research team from industry and academia in Korea has successfully developed an autonomous lab that uses AI and automation to create new cathode materials for secondary batteries. This system operates without human intervention, drastically reducing researcher labor and cutting the material discovery period by 93%.
* Autonomous Lab: A platform that autonomously designs, conducts, and analyzes experiments to find the optimal material.
KAIST (President Kwang Hyung Lee) announced on the 3rd of August that the research team led by Professor Dong-Hwa Seo of the Department of Materials Science and Engineering, in collaboration with the team of LIB Materials Research Center in Energy Materials R&D Laboratories at POSCO Holdings' POSCO N.EX.T Hub (Director Ki Soo Kim), built the lab to explore cathode materials using AI and automation technology.
Developing secondary battery cathode materials is a labor-intensive and time-consuming process for skilled researchers. It involves extensive exploration of various compositions and experimental variables through weighing, transporting, mixing, sintering*, and analyzing samples.
* Sintering: A process in which powder particles are heated to form a single solid mass through thermal activation.
The research team's autonomous lab combines an automated system with an AI model. The system handles all experimental steps—weighing, mixing, pelletizing, sintering, and analysis—without human interference. The AI model then interprets the data, learns from it, and selects the best candidates for the next experiment.
<Figure 1. Outline of the Anode Material Autonomous Exploration Laboratory>
To increase efficiency, the team designed the automation system with separate modules for each process, which are managed by a central robotic arm. This modular approach reduces the system's reliance on the robotic arm.
The team also significantly improved the synthesis speed by using a new high-speed sintering method, which is 50 times faster than the conventional low-speed method. This allows the autonomous lab to acquire 12 times more material data compared to traditional, researcher-led experiments.
<Figure 2. Synthesis of Cathode Material Using a High-Speed Sintering Device>
The vast amount of data collected is automatically interpreted by the AI model to extract information such as synthesized phases and impurity ratios. This data is systematically stored to create a high-quality database, which then serves as training data for an optimization AI model. This creates a closed-loop experimental system that recommends the next cathode composition and synthesis conditions for the automated system.
* Closed-loop experimental system: A system that independently performs all experimental processes without researcher intervention.
Operating this intelligent automation system 24 hours a day can secure more than 12 times the experimental data and shorten material discovery time by 93%. For a project requiring 500 experiments, the system can complete the work in about 6 days, whereas a traditional researcher-led approach would take 84 days.
During development, POSCO Holdings team managed the overall project planning, reviewed the platform design, and co-developed the partial module design and AI-based experimental model. The KAIST team, led by Professor Dong-hwa Seo, was responsible for the actual system implementation and operation, including platform design, module fabrication, algorithm creation, and system verification and improvement.
Professor Dong-Hwa Seo of KAIST stated that this system is a solution to the decrease in research personnel due to the low birth rate in Korea. He expects it will enhance global competitiveness by accelerating secondary battery material development through the acquisition of high-quality data.
<Figure 3. Exterior View (Side) of the Cathode Material Autonomous Exploration Laboratory>
POSCO N.EX.T Hub plans to apply an upgraded version of this autonomous lab to its own research facilities after 2026 to dramatically speed up next-generation secondary battery material development. They are planning further developments to enhance the system's stability and scalability, and hope this industry-academia collaboration will serve as a model for using innovative technology in real-world R&D.
<Figure 4. Exterior View (Front) of the Cathode Material Autonomous Exploration Laboratory>
The research was spearheaded by Ph.D. student Hyun-Gi Lee, along with master's students Seongjae Bae and Dongwoo Kim from Professor Dong-Hwa Seo’s lab at KAIST. Senior researchers Jung Woo Park and Inchul Park from LIB Materials Research Center of POSCO N.EX.T Hub's Energy Materials R&D Laboratories (Director Jeongjin Hong) also participated.
2025.08.06 View 140
Material Innovation Realized with Robotic Arms and AI, Without Human Researchers
<(From Left) M.S candidate Dongwoo Kim from KAIST, Ph.D candidate Hyun-Gi Lee from KAIST, Intern Yeham Kang from KAIST, M.S candidate Seongjae Bae from KAIST, Professor Dong-Hwa Seo from KAIST, (From top right, from left) Senior Researcher Inchul Park from POSCO Holdings, Senior Researcher Jung Woo Park, senior researcher from POSCO Holdings>
A joint research team from industry and academia in Korea has successfully developed an autonomous lab that uses AI and automation to create new cathode materials for secondary batteries. This system operates without human intervention, drastically reducing researcher labor and cutting the material discovery period by 93%.
* Autonomous Lab: A platform that autonomously designs, conducts, and analyzes experiments to find the optimal material.
KAIST (President Kwang Hyung Lee) announced on the 3rd of August that the research team led by Professor Dong-Hwa Seo of the Department of Materials Science and Engineering, in collaboration with the team of LIB Materials Research Center in Energy Materials R&D Laboratories at POSCO Holdings' POSCO N.EX.T Hub (Director Ki Soo Kim), built the lab to explore cathode materials using AI and automation technology.
Developing secondary battery cathode materials is a labor-intensive and time-consuming process for skilled researchers. It involves extensive exploration of various compositions and experimental variables through weighing, transporting, mixing, sintering*, and analyzing samples.
* Sintering: A process in which powder particles are heated to form a single solid mass through thermal activation.
The research team's autonomous lab combines an automated system with an AI model. The system handles all experimental steps—weighing, mixing, pelletizing, sintering, and analysis—without human interference. The AI model then interprets the data, learns from it, and selects the best candidates for the next experiment.
<Figure 1. Outline of the Anode Material Autonomous Exploration Laboratory>
To increase efficiency, the team designed the automation system with separate modules for each process, which are managed by a central robotic arm. This modular approach reduces the system's reliance on the robotic arm.
The team also significantly improved the synthesis speed by using a new high-speed sintering method, which is 50 times faster than the conventional low-speed method. This allows the autonomous lab to acquire 12 times more material data compared to traditional, researcher-led experiments.
<Figure 2. Synthesis of Cathode Material Using a High-Speed Sintering Device>
The vast amount of data collected is automatically interpreted by the AI model to extract information such as synthesized phases and impurity ratios. This data is systematically stored to create a high-quality database, which then serves as training data for an optimization AI model. This creates a closed-loop experimental system that recommends the next cathode composition and synthesis conditions for the automated system.
* Closed-loop experimental system: A system that independently performs all experimental processes without researcher intervention.
Operating this intelligent automation system 24 hours a day can secure more than 12 times the experimental data and shorten material discovery time by 93%. For a project requiring 500 experiments, the system can complete the work in about 6 days, whereas a traditional researcher-led approach would take 84 days.
During development, POSCO Holdings team managed the overall project planning, reviewed the platform design, and co-developed the partial module design and AI-based experimental model. The KAIST team, led by Professor Dong-hwa Seo, was responsible for the actual system implementation and operation, including platform design, module fabrication, algorithm creation, and system verification and improvement.
Professor Dong-Hwa Seo of KAIST stated that this system is a solution to the decrease in research personnel due to the low birth rate in Korea. He expects it will enhance global competitiveness by accelerating secondary battery material development through the acquisition of high-quality data.
<Figure 3. Exterior View (Side) of the Cathode Material Autonomous Exploration Laboratory>
POSCO N.EX.T Hub plans to apply an upgraded version of this autonomous lab to its own research facilities after 2026 to dramatically speed up next-generation secondary battery material development. They are planning further developments to enhance the system's stability and scalability, and hope this industry-academia collaboration will serve as a model for using innovative technology in real-world R&D.
<Figure 4. Exterior View (Front) of the Cathode Material Autonomous Exploration Laboratory>
The research was spearheaded by Ph.D. student Hyun-Gi Lee, along with master's students Seongjae Bae and Dongwoo Kim from Professor Dong-Hwa Seo’s lab at KAIST. Senior researchers Jung Woo Park and Inchul Park from LIB Materials Research Center of POSCO N.EX.T Hub's Energy Materials R&D Laboratories (Director Jeongjin Hong) also participated.
2025.08.06 View 140 -
 Immune Signals Directly Modulate Brain's Emotional Circuits: Unraveling the Mechanism Behind Anxiety-Inducing Behaviors
KAIST's Department of Brain and Cognitive Sciences, led by Professor Jeong-Tae Kwon, has collaborated with MIT and Harvard Medical School to make a groundbreaking discovery. For the first time globally, their joint research has revealed that cytokines, released during immune responses, directly influence the brain's emotional circuits to regulate anxiety behavior.
The study provided experimental evidence for a bidirectional regulatory mechanism: inflammatory cytokines IL-17A and IL-17C act on specific neurons in the amygdala, a region known for emotional regulation, increasing their excitability and consequently inducing anxiety. Conversely, the anti-inflammatory cytokine IL-10 was found to suppress excitability in these very same neurons, thereby contributing to anxiety alleviation.
In a mouse model, the research team observed that while skin inflammation was mitigated by immunotherapy (IL-17RA antibody), anxiety levels paradoxically rose. This was attributed to elevated circulating IL-17 family cytokines leading to the overactivation of amygdala neurons.
Key finding: Inflammatory cytokines IL-17A/17C promote anxiety by acting on excitable amygdala neurons (via IL-17RA/RE receptors), whereas anti-inflammatory cytokine IL-10 alleviates anxiety by suppressing excitability through IL-10RA receptors on the same neurons.
The researchers further elucidated that the anti-inflammatory cytokine IL-10 works to reduce the excitability of these amygdala neurons, thereby mitigating anxiety responses.
This research marks the first instance of demonstrating that immune responses, such as infections or inflammation, directly impact emotional regulation at the level of brain circuits, extending beyond simple physical reactions. This is a profoundly significant achievement, as it proposes a crucial biological mechanism that interlinks immunity, emotion, and behavior through identical neurons within the brain.
The findings of this research were published in the esteemed international journal Cell on April 17th of this year.
Paper Information:
Title: Inflammatory and anti-inflammatory cytokines bidirectionally modulate amygdala circuits regulating anxiety
Journal: Cell (Vol. 188, 2190–2220), April 17, 2025
DOI: https://doi.org/10.1016/j.cell.2025.03.005
Corresponding Authors: Professor Gloria Choi (MIT), Professor Jun R. Huh (Harvard Medical School)
2025.07.24 View 424
Immune Signals Directly Modulate Brain's Emotional Circuits: Unraveling the Mechanism Behind Anxiety-Inducing Behaviors
KAIST's Department of Brain and Cognitive Sciences, led by Professor Jeong-Tae Kwon, has collaborated with MIT and Harvard Medical School to make a groundbreaking discovery. For the first time globally, their joint research has revealed that cytokines, released during immune responses, directly influence the brain's emotional circuits to regulate anxiety behavior.
The study provided experimental evidence for a bidirectional regulatory mechanism: inflammatory cytokines IL-17A and IL-17C act on specific neurons in the amygdala, a region known for emotional regulation, increasing their excitability and consequently inducing anxiety. Conversely, the anti-inflammatory cytokine IL-10 was found to suppress excitability in these very same neurons, thereby contributing to anxiety alleviation.
In a mouse model, the research team observed that while skin inflammation was mitigated by immunotherapy (IL-17RA antibody), anxiety levels paradoxically rose. This was attributed to elevated circulating IL-17 family cytokines leading to the overactivation of amygdala neurons.
Key finding: Inflammatory cytokines IL-17A/17C promote anxiety by acting on excitable amygdala neurons (via IL-17RA/RE receptors), whereas anti-inflammatory cytokine IL-10 alleviates anxiety by suppressing excitability through IL-10RA receptors on the same neurons.
The researchers further elucidated that the anti-inflammatory cytokine IL-10 works to reduce the excitability of these amygdala neurons, thereby mitigating anxiety responses.
This research marks the first instance of demonstrating that immune responses, such as infections or inflammation, directly impact emotional regulation at the level of brain circuits, extending beyond simple physical reactions. This is a profoundly significant achievement, as it proposes a crucial biological mechanism that interlinks immunity, emotion, and behavior through identical neurons within the brain.
The findings of this research were published in the esteemed international journal Cell on April 17th of this year.
Paper Information:
Title: Inflammatory and anti-inflammatory cytokines bidirectionally modulate amygdala circuits regulating anxiety
Journal: Cell (Vol. 188, 2190–2220), April 17, 2025
DOI: https://doi.org/10.1016/j.cell.2025.03.005
Corresponding Authors: Professor Gloria Choi (MIT), Professor Jun R. Huh (Harvard Medical School)
2025.07.24 View 424 -
 Approaches to Human-Robot Interaction Using Biosignals
<(From left) Dr. Hwa-young Jeong, Professor Kyung-seo Park, Dr. Yoon-tae Jeong, Dr. Ji-hoon Seo, Professor Min-kyu Je, Professor Jung Kim >
A joint research team led by Professor Jung Kim of KAIST Department of Mechanical Engineering and Professor Min-kyu Je of the Department of Electrical and Electronic Engineering recently published a review paper on the latest trends and advancements in intuitive Human-Robot Interaction (HRI) using bio-potential and bio-impedance in the internationally renowned academic journal 'Nature Reviews Electrical Engineering'.
This review paper is the result of a collaborative effort by Dr. Kyung-seo Park (DGIST, co-first author), Dr. Hwa-young Jeong (EPFL, co-first author), Dr. Yoon-tae Jeong (IMEC), and Dr. Ji-hoon Seo (UCSD), all doctoral graduates from the two laboratories. Nature Reviews Electrical Engineering is a review specialized journal in the field of electrical, electronic, and artificial intelligence technology, newly launched by Nature Publishing Group last year. It is known to invite world-renowned scholars in the field through strict selection criteria. Professor Jung Kim's research team's paper, titled "Using bio-potential and bio-impedance for intuitive human-robot interaction," was published on July 18, 2025. (DOI: https://doi.org/10.1038/s44287-025-00191-5)
This review paper explains how biosignals can be used to quickly and accurately detect movement intentions and introduces advancements in movement prediction technology based on neural signals and muscle activity. It also focuses on the crucial role of integrated circuits (ICs) in maximizing low-noise performance and energy efficiency in biosignal sensing, covering thelatest development trends in low-noise, low-power designs for accurately measuring bio-potential and impedance signals.
The review emphasizes the importance of hybrid and multi-modal sensing approaches, presenting the possibility of building robust, intuitive, and scalable HRI systems. The research team stressed that collaboration between sensor and IC design fields is essential for the practical application of biosignal-based HRI systems and stated that interdisciplinary collaboration will play a significant role in the development of next-generation HRI technology. Dr. Hwa-young Jeong, a co-first author of the paper, presented the potential of bio-potential and impedance signals to make human-robot interaction more intuitive and efficient, predicting that it will make significant contributions to the development of HRI technologies such as rehabilitation robots and robotic prostheses using biosignals in the future. This research was supported by several research projects, including the Human Plus Project of the National Research Foundation of Korea.
2025.07.24 View 466
Approaches to Human-Robot Interaction Using Biosignals
<(From left) Dr. Hwa-young Jeong, Professor Kyung-seo Park, Dr. Yoon-tae Jeong, Dr. Ji-hoon Seo, Professor Min-kyu Je, Professor Jung Kim >
A joint research team led by Professor Jung Kim of KAIST Department of Mechanical Engineering and Professor Min-kyu Je of the Department of Electrical and Electronic Engineering recently published a review paper on the latest trends and advancements in intuitive Human-Robot Interaction (HRI) using bio-potential and bio-impedance in the internationally renowned academic journal 'Nature Reviews Electrical Engineering'.
This review paper is the result of a collaborative effort by Dr. Kyung-seo Park (DGIST, co-first author), Dr. Hwa-young Jeong (EPFL, co-first author), Dr. Yoon-tae Jeong (IMEC), and Dr. Ji-hoon Seo (UCSD), all doctoral graduates from the two laboratories. Nature Reviews Electrical Engineering is a review specialized journal in the field of electrical, electronic, and artificial intelligence technology, newly launched by Nature Publishing Group last year. It is known to invite world-renowned scholars in the field through strict selection criteria. Professor Jung Kim's research team's paper, titled "Using bio-potential and bio-impedance for intuitive human-robot interaction," was published on July 18, 2025. (DOI: https://doi.org/10.1038/s44287-025-00191-5)
This review paper explains how biosignals can be used to quickly and accurately detect movement intentions and introduces advancements in movement prediction technology based on neural signals and muscle activity. It also focuses on the crucial role of integrated circuits (ICs) in maximizing low-noise performance and energy efficiency in biosignal sensing, covering thelatest development trends in low-noise, low-power designs for accurately measuring bio-potential and impedance signals.
The review emphasizes the importance of hybrid and multi-modal sensing approaches, presenting the possibility of building robust, intuitive, and scalable HRI systems. The research team stressed that collaboration between sensor and IC design fields is essential for the practical application of biosignal-based HRI systems and stated that interdisciplinary collaboration will play a significant role in the development of next-generation HRI technology. Dr. Hwa-young Jeong, a co-first author of the paper, presented the potential of bio-potential and impedance signals to make human-robot interaction more intuitive and efficient, predicting that it will make significant contributions to the development of HRI technologies such as rehabilitation robots and robotic prostheses using biosignals in the future. This research was supported by several research projects, including the Human Plus Project of the National Research Foundation of Korea.
2025.07.24 View 466 -
 KAIST Team Develops Optogenetic Platform for Spatiotemporal Control of Protein and mRNA Storage and Release
<Dr. Chaeyeon Lee, Professor Won Do Heo from Department of Biological Sciences>
A KAIST research team led by Professor Won Do Heo (Department of Biological Sciences) has developed an optogenetic platform, RELISR (REversible LIght-induced Store and Release), that enables precise spatiotemporal control over the storage and release of proteins and mRNAs in living cells and animals.
Traditional optogenetic condensate systems have been limited by their reliance on non-specific multivalent interactions, which can lead to unintended sequestration or release of endogenous molecules. RELISR overcomes these limitations by employing highly specific protein–protein (nanobody–antigen) and protein–RNA (MCP–MS2) interactions, enabling the selective and reversible compartmentalization of target proteins or mRNAs within engineered, membrane-less condensates.
In the dark, RELISR stably sequesters target molecules within condensates, physically isolating them from the cellular environment. Upon blue light stimulation, the condensates rapidly dissolve, releasing the stored proteins or mRNAs, which immediately regain their cellular functions or translational competency. This allows for reversible and rapid modulation of molecular activities in response to optical cues.
< Figure 1. Overview of the Artificial Condensate System (RELISR). The artificial condensate system, RELISR, includes "Protein-RELISR" for storing proteins and "mRNA-RELISR" for storing mRNA. These artificial condensates can be disassembled by blue light irradiation and reassembled in a dark state>
The research team demonstrated that RELISR enables temporal and spatial regulation of protein activity and mRNA translation in various cell types, including cultured neurons and mouse liver tissue. Comparative studies showed that RELISR provides more robust and reversible control of translation than previous systems based on spatial translocation.
While previous optogenetic systems such as LARIAT (Lee et al., Nature Methods, 2014) and mRNA-LARIAT (Kim et al., Nat. Cell Biol., 2019) enabled the selective sequestration of proteins or mRNAs into membrane-less condensates in response to light, they were primarily limited to the trapping phase. The RELISR platform introduced in this study establishes a new paradigm by enabling both the targeted storage of proteins and mRNAs and their rapid, light-triggered release. This approach allows researchers to not only confine molecular function on demand, but also to restore activity with precise temporal control.
< Figure 2. Cell shape change using the artificial condensate system (RELISR). A target protein, Vav2, which contributes to cell shape, was stored within the artificial condensate and then released after light irradiation. This release activated the target protein Vav2, causing a change in cell shape. It was confirmed that the storage, release, and activation of various proteins were effectively achieved>
Professor Heo stated, “RELISR is a versatile optogenetic tool that enables the precise control of protein and mRNA function at defined times and locations in living systems. We anticipate this platform will be broadly applicable for studies of cell signaling, neural circuits, and therapeutic development. Furthermore, the combination of RELISR with genome editing or tissue-targeted delivery could further expand its utility for molecular medicine.”
< Figure 3. Expression of a target mRNA using the artificial condensate system (RELISR) in mice. The genetic material for the artificial condensate system, RELISR, was injected into a living mouse. Using this system, a target mRNA was stored within the mouse's liver. Upon light irradiation, the mRNA was released, which induced the translation of a luminescent protein>
This research was conducted by first author Dr. Chaeyeon Lee, under the supervision of Professor Heo, with contributions from Dr. Daseuli Yu (co-corresponding author) and Professor YongKeun Park (co-corresponding author, Department of Physics), whose group performed quantitative imaging analyses of biophysical changes induced by RELISR in cells.
The findings were published in Nature Communications (July 7, 2025; DOI: 10.1038/s41467-025-61322-y). This work was supported by the Samsung Future Technology Foundation and the National Research Foundation of Korea.
2025.07.23 View 306
KAIST Team Develops Optogenetic Platform for Spatiotemporal Control of Protein and mRNA Storage and Release
<Dr. Chaeyeon Lee, Professor Won Do Heo from Department of Biological Sciences>
A KAIST research team led by Professor Won Do Heo (Department of Biological Sciences) has developed an optogenetic platform, RELISR (REversible LIght-induced Store and Release), that enables precise spatiotemporal control over the storage and release of proteins and mRNAs in living cells and animals.
Traditional optogenetic condensate systems have been limited by their reliance on non-specific multivalent interactions, which can lead to unintended sequestration or release of endogenous molecules. RELISR overcomes these limitations by employing highly specific protein–protein (nanobody–antigen) and protein–RNA (MCP–MS2) interactions, enabling the selective and reversible compartmentalization of target proteins or mRNAs within engineered, membrane-less condensates.
In the dark, RELISR stably sequesters target molecules within condensates, physically isolating them from the cellular environment. Upon blue light stimulation, the condensates rapidly dissolve, releasing the stored proteins or mRNAs, which immediately regain their cellular functions or translational competency. This allows for reversible and rapid modulation of molecular activities in response to optical cues.
< Figure 1. Overview of the Artificial Condensate System (RELISR). The artificial condensate system, RELISR, includes "Protein-RELISR" for storing proteins and "mRNA-RELISR" for storing mRNA. These artificial condensates can be disassembled by blue light irradiation and reassembled in a dark state>
The research team demonstrated that RELISR enables temporal and spatial regulation of protein activity and mRNA translation in various cell types, including cultured neurons and mouse liver tissue. Comparative studies showed that RELISR provides more robust and reversible control of translation than previous systems based on spatial translocation.
While previous optogenetic systems such as LARIAT (Lee et al., Nature Methods, 2014) and mRNA-LARIAT (Kim et al., Nat. Cell Biol., 2019) enabled the selective sequestration of proteins or mRNAs into membrane-less condensates in response to light, they were primarily limited to the trapping phase. The RELISR platform introduced in this study establishes a new paradigm by enabling both the targeted storage of proteins and mRNAs and their rapid, light-triggered release. This approach allows researchers to not only confine molecular function on demand, but also to restore activity with precise temporal control.
< Figure 2. Cell shape change using the artificial condensate system (RELISR). A target protein, Vav2, which contributes to cell shape, was stored within the artificial condensate and then released after light irradiation. This release activated the target protein Vav2, causing a change in cell shape. It was confirmed that the storage, release, and activation of various proteins were effectively achieved>
Professor Heo stated, “RELISR is a versatile optogenetic tool that enables the precise control of protein and mRNA function at defined times and locations in living systems. We anticipate this platform will be broadly applicable for studies of cell signaling, neural circuits, and therapeutic development. Furthermore, the combination of RELISR with genome editing or tissue-targeted delivery could further expand its utility for molecular medicine.”
< Figure 3. Expression of a target mRNA using the artificial condensate system (RELISR) in mice. The genetic material for the artificial condensate system, RELISR, was injected into a living mouse. Using this system, a target mRNA was stored within the mouse's liver. Upon light irradiation, the mRNA was released, which induced the translation of a luminescent protein>
This research was conducted by first author Dr. Chaeyeon Lee, under the supervision of Professor Heo, with contributions from Dr. Daseuli Yu (co-corresponding author) and Professor YongKeun Park (co-corresponding author, Department of Physics), whose group performed quantitative imaging analyses of biophysical changes induced by RELISR in cells.
The findings were published in Nature Communications (July 7, 2025; DOI: 10.1038/s41467-025-61322-y). This work was supported by the Samsung Future Technology Foundation and the National Research Foundation of Korea.
2025.07.23 View 306 -
 Why Do Plants Attack Themselves? The Secret of Genetic Conflict Revealed
<Professor Ji-Joon Song of the KAIST Department of Biological Sciences>
Plants, with their unique immune systems, sometimes launch 'autoimmune responses' by mistakenly identifying their own protein structures as pathogens. In particular, 'hybrid necrosis,' a phenomenon where descendant plants fail to grow healthily and perish after cross-breeding different varieties, has long been a difficult challenge for botanists and agricultural researchers. In response, an international research team has successfully elucidated the mechanism inducing plant autoimmune responses and proposed a novel strategy for cultivar improvement that can predict and avoid these reactions.
Professor Ji-Joon Song's research team at KAIST, in collaboration with teams from the National University of Singapore (NUS) and the University of Oxford, announced on the 21st of July that they have elucidated the structure and function of the 'DM3' protein complex, which triggers plant autoimmune responses, using cryo-electron microscopy (Cryo-EM) technology.
This research is drawing attention because it identifies defects in protein structure as the cause of hybrid necrosis, which occurs due to an abnormal reaction of immune receptors during cross-breeding between plant hybrids.
This protein (DM3) is originally an enzyme involved in the plant's immune response, but problems arise when the structure of the DM3 protein is damaged in a specific protein combination called 'DANGEROUS MIX (DM)'.
Notably, one variant of DM3, the 'DM3Col-0' variant, forms a stable complex with six proteins and is recognized as normal, thus not triggering an immune response. In contrast, another 'DM3Hh-0' variant has improper binding between its six proteins, causing the plant to recognize it as an 'abnormal state' and trigger an immune alarm, leading to autoimmunity.
The research team visualized this structure using atomic-resolution cryo-electron microscopy (Cryo-EM) and revealed that the immune-inducing ability is not due to the enzymatic function of the DM3 protein, but rather to 'differences in protein binding affinity.'
<Figure 1. Mechanism of Plant Autoimmunity Triggered by the Collapse of the DM3 Protein Complex>
This demonstrates that plants can initiate an immune response by recognizing not only 'external pathogens' but also 'internal protein structures' when they undergo abnormal changes, treating them as if they were pathogens.
The study shows how sensitively the plant immune system changes and triggers autoimmune responses when genes are mixed and protein structures change during the cross-breeding of different plant varieties. It significantly advanced the understanding of genetic incompatibility that can occur during natural cross-breeding and cultivar improvement processes.
Dr. Gijeong Kim, the co-first author, stated, "Through international research collaboration, we presented a new perspective on understanding the plant immune system by leveraging the autoimmune phenomenon, completing a high-quality study that encompasses structural biochemistry, genetics, and cell biological experiments."
Professor Ji-Joon Song of the KAIST Department of Biological Sciences, who led the research, said, "The fact that the immune system can detect not only external pathogens but also structural abnormalities in its own proteins will set a new standard for plant biotechnology and crop breeding strategies. Cryo-electron microscopy-based structural analysis will be an important tool for understanding the essence of gene interactions."
This research, with Professor Ji-Joon Song and Professor Eunyoung Chae of the University of Oxford as co-corresponding authors, Dr. Gijeong Kim (currently a postdoctoral researcher at the University of Zurich) and Dr. Wei-Lin Wan of the National University of Singapore as co-first authors, and Ph.D candidate Nayun Kim, as the second author, was published on July 17th in Molecular Cell, a sister journal of the international academic journal Cell.
This research was supported by the KAIST Grand Challenge 30 project.
Article Title: Structural determinants of DANGEROUS MIX 3, an alpha/beta hydrolase that triggers NLR-mediated genetic incompatibility in plants DOI: https://doi.org/10.1016/j.molcel.2025.06.021
2025.07.21 View 467
Why Do Plants Attack Themselves? The Secret of Genetic Conflict Revealed
<Professor Ji-Joon Song of the KAIST Department of Biological Sciences>
Plants, with their unique immune systems, sometimes launch 'autoimmune responses' by mistakenly identifying their own protein structures as pathogens. In particular, 'hybrid necrosis,' a phenomenon where descendant plants fail to grow healthily and perish after cross-breeding different varieties, has long been a difficult challenge for botanists and agricultural researchers. In response, an international research team has successfully elucidated the mechanism inducing plant autoimmune responses and proposed a novel strategy for cultivar improvement that can predict and avoid these reactions.
Professor Ji-Joon Song's research team at KAIST, in collaboration with teams from the National University of Singapore (NUS) and the University of Oxford, announced on the 21st of July that they have elucidated the structure and function of the 'DM3' protein complex, which triggers plant autoimmune responses, using cryo-electron microscopy (Cryo-EM) technology.
This research is drawing attention because it identifies defects in protein structure as the cause of hybrid necrosis, which occurs due to an abnormal reaction of immune receptors during cross-breeding between plant hybrids.
This protein (DM3) is originally an enzyme involved in the plant's immune response, but problems arise when the structure of the DM3 protein is damaged in a specific protein combination called 'DANGEROUS MIX (DM)'.
Notably, one variant of DM3, the 'DM3Col-0' variant, forms a stable complex with six proteins and is recognized as normal, thus not triggering an immune response. In contrast, another 'DM3Hh-0' variant has improper binding between its six proteins, causing the plant to recognize it as an 'abnormal state' and trigger an immune alarm, leading to autoimmunity.
The research team visualized this structure using atomic-resolution cryo-electron microscopy (Cryo-EM) and revealed that the immune-inducing ability is not due to the enzymatic function of the DM3 protein, but rather to 'differences in protein binding affinity.'
<Figure 1. Mechanism of Plant Autoimmunity Triggered by the Collapse of the DM3 Protein Complex>
This demonstrates that plants can initiate an immune response by recognizing not only 'external pathogens' but also 'internal protein structures' when they undergo abnormal changes, treating them as if they were pathogens.
The study shows how sensitively the plant immune system changes and triggers autoimmune responses when genes are mixed and protein structures change during the cross-breeding of different plant varieties. It significantly advanced the understanding of genetic incompatibility that can occur during natural cross-breeding and cultivar improvement processes.
Dr. Gijeong Kim, the co-first author, stated, "Through international research collaboration, we presented a new perspective on understanding the plant immune system by leveraging the autoimmune phenomenon, completing a high-quality study that encompasses structural biochemistry, genetics, and cell biological experiments."
Professor Ji-Joon Song of the KAIST Department of Biological Sciences, who led the research, said, "The fact that the immune system can detect not only external pathogens but also structural abnormalities in its own proteins will set a new standard for plant biotechnology and crop breeding strategies. Cryo-electron microscopy-based structural analysis will be an important tool for understanding the essence of gene interactions."
This research, with Professor Ji-Joon Song and Professor Eunyoung Chae of the University of Oxford as co-corresponding authors, Dr. Gijeong Kim (currently a postdoctoral researcher at the University of Zurich) and Dr. Wei-Lin Wan of the National University of Singapore as co-first authors, and Ph.D candidate Nayun Kim, as the second author, was published on July 17th in Molecular Cell, a sister journal of the international academic journal Cell.
This research was supported by the KAIST Grand Challenge 30 project.
Article Title: Structural determinants of DANGEROUS MIX 3, an alpha/beta hydrolase that triggers NLR-mediated genetic incompatibility in plants DOI: https://doi.org/10.1016/j.molcel.2025.06.021
2025.07.21 View 467 -
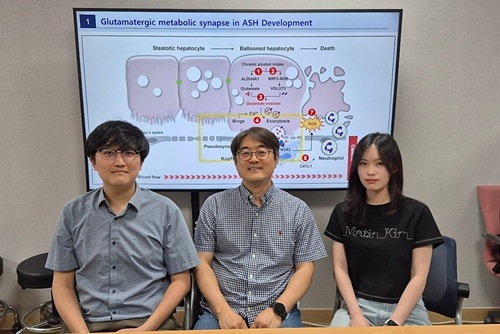 KAIST reveals for the first time the mechanism by which alcohol triggers liver inflammation
<(From left)Dr. Keungmo Yang, Professor Won-Il Jeong, Ph.D candidate Kyurae Kim>
Excessive alcohol consumption causes alcoholic liver disease, and about 20% of these cases progress to alcohol-associated steatohepatitis (ASH), which can lead to liver cirrhosis and liver failure. Early diagnosis and treatment are therefore extremely important. A KAIST research team has identified a new molecular mechanism in which alcohol-damaged liver cells increase reactive oxygen species (ROS), leading to cell death and inflammatory responses. In addition, they discovered that Kupffer cells, immune cells residing in the liver, act as a “dual-function regulator” that can either promote or suppress inflammation through interactions with liver cells.
KAIST (President Kwang-Hyung Lee) announced on the 17th that a research team led by Professor Won-Il Jeong from the Graduate School of Medical Science and Engineering, in collaboration with Professor Won Kim’s team at Seoul National University Boramae Medical Center, has uncovered the molecular pathway of liver damage and inflammation caused by alcohol consumption. This finding offers new clues for the diagnosis and treatment of alcohol-associated liver disease (ALD).
Professor Won-Il Jeong’s research team found that during chronic alcohol intake, expression of the vesicular glutamate transporter VGLUT3 increases, leading to glutamate accumulation in hepatocytes. Subsequent binge drinking causes rapid changes in intracellular calcium levels, which then triggers glutamate* secretion. The secreted glutamate stimulates the glutamate receptor mGluR5 on liver-resident macrophages (Kupffer cells), which induces ROS production and activates a pathological pathway resulting in hepatocyte death and inflammation.
*Glutamate: A type of amino acid involved in intercellular signaling, protein synthesis, and energy metabolism in various tissues including the brain and liver. In excess, it can cause overexcitation and death of nerve cells.
Glutamate accumulation in perivenous hepatocytes through vesicular glutamate transporter 3 after 2-week EtOH intake and its release by binge drinking>
A particularly groundbreaking aspect of this study is that damaged hepatocytes and Kupffer cells can form a "pseudosynapse"—a structure similar to a synapse which is previously thought to occur only in the brain—enabling them to exchange signals. This is the first time such a phenomenon has been identified in the liver.
This pseudosynapse forms when hepatocytes expand (ballooning) due to alcohol, becoming physically attached to Kupffer cells. Simply put, the damaged hepatocytes don’t just die—they send distress signals to nearby immune cells, prompting a response.
This discovery proposes a new paradigm: even in peripheral organs, direct structural contact between cells can allow signal transmission. It also shows that damaged hepatocytes can actively stimulate macrophages and induce regeneration through cell death, revealing the liver’s “autonomous recovery function.”
The team also confirmed in animal models that genetic or pharmacological inhibition of VGLUT3, mGluR5, or the ROS-producing enzyme NOX2 reduces alcohol-induced liver damage. They also confirmed that the same mechanism observed in animal models was present in human patients with ALD by analyzing blood and liver tissue samples.
Professor Won-Il Jeong of KAIST said, “These findings may serve as new molecular targets for early diagnosis and treatment of ASH in the future.”
This study was jointly led by Dr. Keungmo Yang (now at Yeouido St. Mary’s Hospital) and Kyurae Kim, a doctoral candidate at KAIST, who served as co–first authors. It was conducted in collaboration with Professor Won Kim’s team at Seoul National University Boramae Medical Center and was published in the journal Nature Communications on July 1.
※ Article Title: Binge drinking triggers VGLUT3-mediated glutamate secretion and subsequent hepatic inflammation by activating mGluR5/NOX2 in Kupffer cells ※ DOI: https://doi.org/10.1038/s41467-025-60820-3
This study was supported by the Ministry of Science and ICT through the National Research Foundation of Korea's Global Leader Program, Mid-Career Researcher Program, and the Bio & Medical Technology Development Program.
2025.07.17 View 608
KAIST reveals for the first time the mechanism by which alcohol triggers liver inflammation
<(From left)Dr. Keungmo Yang, Professor Won-Il Jeong, Ph.D candidate Kyurae Kim>
Excessive alcohol consumption causes alcoholic liver disease, and about 20% of these cases progress to alcohol-associated steatohepatitis (ASH), which can lead to liver cirrhosis and liver failure. Early diagnosis and treatment are therefore extremely important. A KAIST research team has identified a new molecular mechanism in which alcohol-damaged liver cells increase reactive oxygen species (ROS), leading to cell death and inflammatory responses. In addition, they discovered that Kupffer cells, immune cells residing in the liver, act as a “dual-function regulator” that can either promote or suppress inflammation through interactions with liver cells.
KAIST (President Kwang-Hyung Lee) announced on the 17th that a research team led by Professor Won-Il Jeong from the Graduate School of Medical Science and Engineering, in collaboration with Professor Won Kim’s team at Seoul National University Boramae Medical Center, has uncovered the molecular pathway of liver damage and inflammation caused by alcohol consumption. This finding offers new clues for the diagnosis and treatment of alcohol-associated liver disease (ALD).
Professor Won-Il Jeong’s research team found that during chronic alcohol intake, expression of the vesicular glutamate transporter VGLUT3 increases, leading to glutamate accumulation in hepatocytes. Subsequent binge drinking causes rapid changes in intracellular calcium levels, which then triggers glutamate* secretion. The secreted glutamate stimulates the glutamate receptor mGluR5 on liver-resident macrophages (Kupffer cells), which induces ROS production and activates a pathological pathway resulting in hepatocyte death and inflammation.
*Glutamate: A type of amino acid involved in intercellular signaling, protein synthesis, and energy metabolism in various tissues including the brain and liver. In excess, it can cause overexcitation and death of nerve cells.
Glutamate accumulation in perivenous hepatocytes through vesicular glutamate transporter 3 after 2-week EtOH intake and its release by binge drinking>
A particularly groundbreaking aspect of this study is that damaged hepatocytes and Kupffer cells can form a "pseudosynapse"—a structure similar to a synapse which is previously thought to occur only in the brain—enabling them to exchange signals. This is the first time such a phenomenon has been identified in the liver.
This pseudosynapse forms when hepatocytes expand (ballooning) due to alcohol, becoming physically attached to Kupffer cells. Simply put, the damaged hepatocytes don’t just die—they send distress signals to nearby immune cells, prompting a response.
This discovery proposes a new paradigm: even in peripheral organs, direct structural contact between cells can allow signal transmission. It also shows that damaged hepatocytes can actively stimulate macrophages and induce regeneration through cell death, revealing the liver’s “autonomous recovery function.”
The team also confirmed in animal models that genetic or pharmacological inhibition of VGLUT3, mGluR5, or the ROS-producing enzyme NOX2 reduces alcohol-induced liver damage. They also confirmed that the same mechanism observed in animal models was present in human patients with ALD by analyzing blood and liver tissue samples.
Professor Won-Il Jeong of KAIST said, “These findings may serve as new molecular targets for early diagnosis and treatment of ASH in the future.”
This study was jointly led by Dr. Keungmo Yang (now at Yeouido St. Mary’s Hospital) and Kyurae Kim, a doctoral candidate at KAIST, who served as co–first authors. It was conducted in collaboration with Professor Won Kim’s team at Seoul National University Boramae Medical Center and was published in the journal Nature Communications on July 1.
※ Article Title: Binge drinking triggers VGLUT3-mediated glutamate secretion and subsequent hepatic inflammation by activating mGluR5/NOX2 in Kupffer cells ※ DOI: https://doi.org/10.1038/s41467-025-60820-3
This study was supported by the Ministry of Science and ICT through the National Research Foundation of Korea's Global Leader Program, Mid-Career Researcher Program, and the Bio & Medical Technology Development Program.
2025.07.17 View 608 -
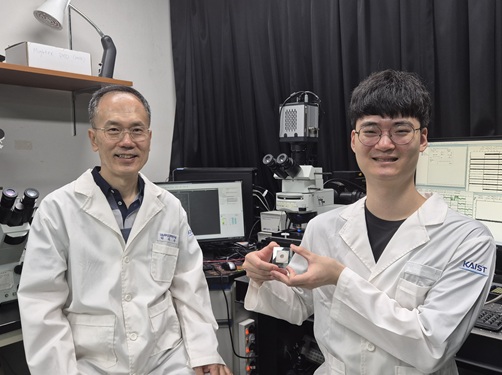 KAIST Successfully Implements 3D Brain-Mimicking Platform with 6x Higher Precision
<(From left) Dr. Dongjo Yoon, Professor Je-Kyun Park from the Department of Bio and Brain Engineering, (upper right) Professor Yoonkey Nam, Dr. Soo Jee Kim>
Existing three-dimensional (3D) neuronal culture technology has limitations in brain research due to the difficulty of precisely replicating the brain's complex multilayered structure and the lack of a platform that can simultaneously analyze both structure and function. A KAIST research team has successfully developed an integrated platform that can implement brain-like layered neuronal structures using 3D printing technology and precisely measure neuronal activity within them.
KAIST (President Kwang Hyung Lee) announced on the 16th of July that a joint research team led by Professors Je-Kyun Park and Yoonkey Nam from the Department of Bio and Brain Engineering has developed an integrated platform capable of fabricating high-resolution 3D multilayer neuronal networks using low-viscosity natural hydrogels with mechanical properties similar to brain tissue, and simultaneously analyzing their structural and functional connectivity.
Conventional bioprinting technology uses high-viscosity bioinks for structural stability, but this limits neuronal proliferation and neurite growth. Conversely, neural cell-friendly low-viscosity hydrogels are difficult to precisely pattern, leading to a fundamental trade-off between structural stability and biological function. The research team completed a sophisticated and stable brain-mimicking platform by combining three key technologies that enable the precise creation of brain structure with dilute gels, accurate alignment between layers, and simultaneous observation of neuronal activity.
The three core technologies are: ▲ 'Capillary Pinning Effect' technology, which enables the dilute gel (hydrogel) to adhere firmly to a stainless steel mesh (micromesh) to prevent it from flowing, thereby reproducing brain structures with six times greater precision (resolution of 500 μm or less) than conventional methods; ▲ the '3D Printing Aligner,' a cylindrical design that ensures the printed layers are precisely stacked without misalignment, guaranteeing the accurate assembly of multilayer structures and stable integration with microelectrode chips; and ▲ 'Dual-mode Analysis System' technology, which simultaneously measures electrical signals from below and observes cell activity with light (calcium imaging) from above, allowing for the simultaneous verification of the functional operation of interlayer connections through multiple methods.
< Figure 1. Platform integrating brain-structure-mimicking neural network model construction and functional measurement technology>
The research team successfully implemented a three-layered mini-brain structure using 3D printing with a fibrin hydrogel, which has elastic properties similar to those of the brain, and experimentally verified the process of actual neural cells transmitting and receiving signals within it.
Cortical neurons were placed in the upper and lower layers, while the middle layer was left empty but designed to allow neurons to penetrate and connect through it. Electrical signals were measured from the lower layer using a microsensor (electrode chip), and cell activity was observed from the upper layer using light (calcium imaging). The results showed that when electrical stimulation was applied, neural cells in both upper and lower layers responded simultaneously. When a synapse-blocking agent (synaptic blocker) was introduced, the response decreased, proving that the neural cells were genuinely connected and transmitting signals.
Professor Je-Kyun Park of KAIST explained, "This research is a joint development achievement of an integrated platform that can simultaneously reproduce the complex multilayered structure and function of brain tissue. Compared to existing technologies where signal measurement was impossible for more than 14 days, this platform maintains a stable microelectrode chip interface for over 27 days, allowing the real-time analysis of structure-function relationships. It can be utilized in various brain research fields such as neurological disease modeling, brain function research, neurotoxicity assessment, and neuroprotective drug screening in the future."
The research, in which Dr. Soo Jee Kim and Dr. Dongjo Yoon from KAIST's Department of Bio and Brain Engineering participated as co-first authors, was published online in the international journal 'Biosensors and Bioelectronics' on June 11, 2025.
※Paper: Hybrid biofabrication of multilayered 3D neuronal networks with structural and functional interlayer connectivity
※DOI: https://doi.org/10.1016/j.bios.2025.117688
2025.07.16 View 540
KAIST Successfully Implements 3D Brain-Mimicking Platform with 6x Higher Precision
<(From left) Dr. Dongjo Yoon, Professor Je-Kyun Park from the Department of Bio and Brain Engineering, (upper right) Professor Yoonkey Nam, Dr. Soo Jee Kim>
Existing three-dimensional (3D) neuronal culture technology has limitations in brain research due to the difficulty of precisely replicating the brain's complex multilayered structure and the lack of a platform that can simultaneously analyze both structure and function. A KAIST research team has successfully developed an integrated platform that can implement brain-like layered neuronal structures using 3D printing technology and precisely measure neuronal activity within them.
KAIST (President Kwang Hyung Lee) announced on the 16th of July that a joint research team led by Professors Je-Kyun Park and Yoonkey Nam from the Department of Bio and Brain Engineering has developed an integrated platform capable of fabricating high-resolution 3D multilayer neuronal networks using low-viscosity natural hydrogels with mechanical properties similar to brain tissue, and simultaneously analyzing their structural and functional connectivity.
Conventional bioprinting technology uses high-viscosity bioinks for structural stability, but this limits neuronal proliferation and neurite growth. Conversely, neural cell-friendly low-viscosity hydrogels are difficult to precisely pattern, leading to a fundamental trade-off between structural stability and biological function. The research team completed a sophisticated and stable brain-mimicking platform by combining three key technologies that enable the precise creation of brain structure with dilute gels, accurate alignment between layers, and simultaneous observation of neuronal activity.
The three core technologies are: ▲ 'Capillary Pinning Effect' technology, which enables the dilute gel (hydrogel) to adhere firmly to a stainless steel mesh (micromesh) to prevent it from flowing, thereby reproducing brain structures with six times greater precision (resolution of 500 μm or less) than conventional methods; ▲ the '3D Printing Aligner,' a cylindrical design that ensures the printed layers are precisely stacked without misalignment, guaranteeing the accurate assembly of multilayer structures and stable integration with microelectrode chips; and ▲ 'Dual-mode Analysis System' technology, which simultaneously measures electrical signals from below and observes cell activity with light (calcium imaging) from above, allowing for the simultaneous verification of the functional operation of interlayer connections through multiple methods.
< Figure 1. Platform integrating brain-structure-mimicking neural network model construction and functional measurement technology>
The research team successfully implemented a three-layered mini-brain structure using 3D printing with a fibrin hydrogel, which has elastic properties similar to those of the brain, and experimentally verified the process of actual neural cells transmitting and receiving signals within it.
Cortical neurons were placed in the upper and lower layers, while the middle layer was left empty but designed to allow neurons to penetrate and connect through it. Electrical signals were measured from the lower layer using a microsensor (electrode chip), and cell activity was observed from the upper layer using light (calcium imaging). The results showed that when electrical stimulation was applied, neural cells in both upper and lower layers responded simultaneously. When a synapse-blocking agent (synaptic blocker) was introduced, the response decreased, proving that the neural cells were genuinely connected and transmitting signals.
Professor Je-Kyun Park of KAIST explained, "This research is a joint development achievement of an integrated platform that can simultaneously reproduce the complex multilayered structure and function of brain tissue. Compared to existing technologies where signal measurement was impossible for more than 14 days, this platform maintains a stable microelectrode chip interface for over 27 days, allowing the real-time analysis of structure-function relationships. It can be utilized in various brain research fields such as neurological disease modeling, brain function research, neurotoxicity assessment, and neuroprotective drug screening in the future."
The research, in which Dr. Soo Jee Kim and Dr. Dongjo Yoon from KAIST's Department of Bio and Brain Engineering participated as co-first authors, was published online in the international journal 'Biosensors and Bioelectronics' on June 11, 2025.
※Paper: Hybrid biofabrication of multilayered 3D neuronal networks with structural and functional interlayer connectivity
※DOI: https://doi.org/10.1016/j.bios.2025.117688
2025.07.16 View 540 -
 A KAIST Team Engineers a Microbial Platform for Efficient Lutein Production
<(From Left) Ph.D. Candidate Hyunmin Eun, Distinguished Professor Sang Yup Lee, , Dr. Cindy Pricilia Surya Prabowo>
The application of systems metabolic engineering strategies, along with the construction of an electron channeling system, has enabled the first gram-per-liter scale production of lutein from Corynebacterium glutamicum, providing a viable alternative to plant-derived lutein production.
A research group at KAIST has successfully engineered a microbial strain capable of producing lutein at industrially relevant levels. The team, led by Distinguished Professor Sang Yup Lee from the Department of Chemical and Biomolecular Engineering, developed a novel C. glutamicum strain using systems metabolic engineering strategies to overcome the limitations of previous microbial lutein production efforts. This research is expected to be beneficial for the efficient production of other industrially important natural products used in food, pharmaceuticals, and cosmetics.
Lutein is a xanthophyll carotenoid found in egg yolk, fruits, and vegetables, known for its role in protecting our eyes from oxidative stress and reducing the risk of macular degeneration and cataracts. Currently, commercial lutein is predominantly extracted from marigold flowers; however, this approach has several drawbacks, including long cultivation times, high labor costs, and inefficient extraction yields, making it economically unfeasible for large-scale production. These challenges have driven the demand for alternative production methods.
To address these issues, KAIST researchers, including Ph.D. Candidate Hyunmin Eun, Dr. Cindy Pricilia Surya Prabowo, and Distinguished Professor Sang Yup Lee, applied systems metabolic engineering strategies to engineer C. glutamicum, a GRAS (Generally Recognized As Safe) microorganism widely used in industrial fermentation. Unlike Escherichia coli, which was previously explored for microbial lutein production, C. glutamicum lacks endotoxins, making it a safer and more viable option for food and pharmaceutical applications.
The team’s work, entitled “Gram-per-litre scale production of lutein by engineered Corynebacterium,” was published in Nature Synthesis on 04 July , 2025.
This research details the high-level production of lutein using glucose as a renewable carbon source via systems metabolic engineering. The team focused on eliminating metabolic bottlenecks that previously limited microbial lutein synthesis. By employing enzyme scaffold-based electron channeling strategies, the researchers improved metabolic flux towards lutein biosynthesis while minimizing unwanted byproducts.
<Lutein production metabolic pathway engineering>
To enhance productivity, bottleneck enzymes within the metabolic pathway were identified and optimized. It was determined that electron-requiring cytochrome P450 enzymes played a major role in limiting lutein biosynthesis. To overcome this limitation, an electron channeling strategy was implemented, where engineered cytochrome P450 enzymes and their reductase partners were spatially organized on synthetic scaffolds, allowing more efficient electron transfer and significantly increasing lutein production.
The engineered C. glutamicum strain was further optimized in fed-batch fermentation, achieving a record-breaking 1.78 g/L of lutein production within 54 hours, with a content of 19.51 mg/gDCW and a productivity of 32.88 mg/L/h—the highest lutein production performance in any host reported to date. This milestone demonstrates the feasibility of replacing plant-based lutein extraction with microbial fermentation technology.
“We can anticipate that this microbial cell factory-based mass production of lutein will be able to replace the current plant extraction-based process,” said Ph.D. Candidate Hyunmin Eun. He emphasized that the integrated metabolic engineering strategies developed in this study could be broadly applied for the efficient production of other valuable natural products used in pharmaceuticals and nutraceuticals.
<Schematic diagram of microbial-based lutein production platform>
“As maintaining good health in an aging society becomes increasingly important, we expect that the technology and strategies developed here will play pivotal roles in producing other medically and nutritionally significant natural products,” added Distinguished Professor Sang Yup Lee.
This work is supported by the Development of Next-generation Biorefinery Platform Technologies for Leading Bio-based Chemicals Industry project 2022M3J5A1056072 and the Development of Platform Technologies of Microbial Cell Factories for the Next-Generation Biorefineries project 2022M3J5A1056117 from the National Research Foundation supported by the Korean Ministry of Science and ICT.
Source:
Hyunmin Eun (1st), Cindy Pricilia Surya Prabowo (co-1st), and Sang Yup Lee (Corresponding). “Gram-per-litre scale production of lutein by engineered Corynebacterium”. Nature Synthesis (Online published)
For further information:
Sang Yup Lee, Distinguished Professor of Chemical and Biomolecular Engineering, KAIST (leesy@kaist.ac.kr, Tel: +82-42-350-3930)
2025.07.14 View 1138
A KAIST Team Engineers a Microbial Platform for Efficient Lutein Production
<(From Left) Ph.D. Candidate Hyunmin Eun, Distinguished Professor Sang Yup Lee, , Dr. Cindy Pricilia Surya Prabowo>
The application of systems metabolic engineering strategies, along with the construction of an electron channeling system, has enabled the first gram-per-liter scale production of lutein from Corynebacterium glutamicum, providing a viable alternative to plant-derived lutein production.
A research group at KAIST has successfully engineered a microbial strain capable of producing lutein at industrially relevant levels. The team, led by Distinguished Professor Sang Yup Lee from the Department of Chemical and Biomolecular Engineering, developed a novel C. glutamicum strain using systems metabolic engineering strategies to overcome the limitations of previous microbial lutein production efforts. This research is expected to be beneficial for the efficient production of other industrially important natural products used in food, pharmaceuticals, and cosmetics.
Lutein is a xanthophyll carotenoid found in egg yolk, fruits, and vegetables, known for its role in protecting our eyes from oxidative stress and reducing the risk of macular degeneration and cataracts. Currently, commercial lutein is predominantly extracted from marigold flowers; however, this approach has several drawbacks, including long cultivation times, high labor costs, and inefficient extraction yields, making it economically unfeasible for large-scale production. These challenges have driven the demand for alternative production methods.
To address these issues, KAIST researchers, including Ph.D. Candidate Hyunmin Eun, Dr. Cindy Pricilia Surya Prabowo, and Distinguished Professor Sang Yup Lee, applied systems metabolic engineering strategies to engineer C. glutamicum, a GRAS (Generally Recognized As Safe) microorganism widely used in industrial fermentation. Unlike Escherichia coli, which was previously explored for microbial lutein production, C. glutamicum lacks endotoxins, making it a safer and more viable option for food and pharmaceutical applications.
The team’s work, entitled “Gram-per-litre scale production of lutein by engineered Corynebacterium,” was published in Nature Synthesis on 04 July , 2025.
This research details the high-level production of lutein using glucose as a renewable carbon source via systems metabolic engineering. The team focused on eliminating metabolic bottlenecks that previously limited microbial lutein synthesis. By employing enzyme scaffold-based electron channeling strategies, the researchers improved metabolic flux towards lutein biosynthesis while minimizing unwanted byproducts.
<Lutein production metabolic pathway engineering>
To enhance productivity, bottleneck enzymes within the metabolic pathway were identified and optimized. It was determined that electron-requiring cytochrome P450 enzymes played a major role in limiting lutein biosynthesis. To overcome this limitation, an electron channeling strategy was implemented, where engineered cytochrome P450 enzymes and their reductase partners were spatially organized on synthetic scaffolds, allowing more efficient electron transfer and significantly increasing lutein production.
The engineered C. glutamicum strain was further optimized in fed-batch fermentation, achieving a record-breaking 1.78 g/L of lutein production within 54 hours, with a content of 19.51 mg/gDCW and a productivity of 32.88 mg/L/h—the highest lutein production performance in any host reported to date. This milestone demonstrates the feasibility of replacing plant-based lutein extraction with microbial fermentation technology.
“We can anticipate that this microbial cell factory-based mass production of lutein will be able to replace the current plant extraction-based process,” said Ph.D. Candidate Hyunmin Eun. He emphasized that the integrated metabolic engineering strategies developed in this study could be broadly applied for the efficient production of other valuable natural products used in pharmaceuticals and nutraceuticals.
<Schematic diagram of microbial-based lutein production platform>
“As maintaining good health in an aging society becomes increasingly important, we expect that the technology and strategies developed here will play pivotal roles in producing other medically and nutritionally significant natural products,” added Distinguished Professor Sang Yup Lee.
This work is supported by the Development of Next-generation Biorefinery Platform Technologies for Leading Bio-based Chemicals Industry project 2022M3J5A1056072 and the Development of Platform Technologies of Microbial Cell Factories for the Next-Generation Biorefineries project 2022M3J5A1056117 from the National Research Foundation supported by the Korean Ministry of Science and ICT.
Source:
Hyunmin Eun (1st), Cindy Pricilia Surya Prabowo (co-1st), and Sang Yup Lee (Corresponding). “Gram-per-litre scale production of lutein by engineered Corynebacterium”. Nature Synthesis (Online published)
For further information:
Sang Yup Lee, Distinguished Professor of Chemical and Biomolecular Engineering, KAIST (leesy@kaist.ac.kr, Tel: +82-42-350-3930)
2025.07.14 View 1138 -
 KAIST Ushers in Era of Predicting ‘Optimal Alloys’ Using AI, Without High-Temperature Experiments
<Picture1.(From Left) Prof. Seungbum Hong, Ph.D candidate Youngwoo Choi>
Steel alloys used in automobiles and machinery parts are typically manufactured through a melting process at high temperatures. The phenomenon where the components remain unchanged during melting is called “congruent melting.” KAIST researchers have now addressed this process—traditionally only possible through high-temperature experiments—using artificial intelligence (AI). This study draws attention as it proposes a new direction for future alloy development by predicting in advance how well alloy components will mix during melting, a long-standing challenge in the field.
KAIST (President Kwang Hyung Lee) announced on the 14th of July that Professor Seungbum Hong’s research team from the Department of Materials Science and Engineering, in international collaboration with Professor Chris Wolverton’s group at Northwestern University, has developed a high-accuracy machine learning model that predicts whether alloy components will remain stable during melting. This was achieved using formation energy data derived from Density Functional Theory (DFT)* calculations.
*Density Functional Theory (DFT): A computational quantum mechanical method used to investigate the electronic structure of many-body systems, especially atoms, molecules, and solids, based on electron density.
The research team combined formation energy values obtained via DFT with experimental melting reaction data to train a machine learning model on 4,536 binary compounds.
Among the various machine learning algorithms tested, the XGBoost-based classification model demonstrated the highest accuracy in predicting whether alloys would mix well, achieving a prediction accuracy of approximately 82.5%. The team also applied the Shapley value method* to analyze the key features of the model. One major finding was that sharp changes in the slope of the formation energy curve (referred to as “convex hull sharpness”) were the most significant factor. A steep slope indicates a composition with energetically favorable (i.e., stable) formation.
*Shapley value: An explainability method in AI used to determine how much each feature contributed to a prediction.
The most notable significance of this study is that it predicts alloy melting behavior without performing high-temperature experiments. This is especially useful for materials such as high-entropy alloys or ultra-heat-resistant alloys, which are difficult to handle experimentally. The approach could also be extended to the design of complex multi-component alloy systems in the future.
Furthermore, the physical indicators identified by the AI model showed high consistency with actual experimental results on how well alloys mix and remain stable. This suggests that the model could be broadly applied to the development of various metal materials and the prediction of structural stability.
Professor Seungbum Hong of KAIST stated, “This research demonstrates how data-driven predictive materials development is possible by integrating computational methods, experimental data, and machine learning—departing from the traditional experience-based alloy design.” He added, “In the future, by incorporating state-of-the-art AI techniques such as generative models and reinforcement learning, we could enter an era where completely new alloys are designed automatically.”
<Model performance and feature importance analysis for predicting melting congruency. (a) SHAP summary plot showing the impact of individual features on model predictions. (b) Confusion matrix illustrating the model’s classification performance. (c) Receiver operating characteristic (ROC) curve with an AUC (area under the curve) score of 0.87, indicating a strong classification performance.>
Ph.D. candidate Youngwoo Choi, from the Department of Materials Science and Engineering at KAIST, participated as the first author. The study was published in the May issue of APL Machine Learning, a prestigious journal in the field of machine learning published by the American Institute of Physics, and was selected as a “Featured Article.”
※ Paper title: Machine learning-based melting congruency prediction of binary compounds using density functional theory-calculated formation energy ※ DOI: 10.1063/5.0247514
This research was supported by the Ministry of Science and ICT and the National Research Foundation of Korea.
2025.07.14 View 433
KAIST Ushers in Era of Predicting ‘Optimal Alloys’ Using AI, Without High-Temperature Experiments
<Picture1.(From Left) Prof. Seungbum Hong, Ph.D candidate Youngwoo Choi>
Steel alloys used in automobiles and machinery parts are typically manufactured through a melting process at high temperatures. The phenomenon where the components remain unchanged during melting is called “congruent melting.” KAIST researchers have now addressed this process—traditionally only possible through high-temperature experiments—using artificial intelligence (AI). This study draws attention as it proposes a new direction for future alloy development by predicting in advance how well alloy components will mix during melting, a long-standing challenge in the field.
KAIST (President Kwang Hyung Lee) announced on the 14th of July that Professor Seungbum Hong’s research team from the Department of Materials Science and Engineering, in international collaboration with Professor Chris Wolverton’s group at Northwestern University, has developed a high-accuracy machine learning model that predicts whether alloy components will remain stable during melting. This was achieved using formation energy data derived from Density Functional Theory (DFT)* calculations.
*Density Functional Theory (DFT): A computational quantum mechanical method used to investigate the electronic structure of many-body systems, especially atoms, molecules, and solids, based on electron density.
The research team combined formation energy values obtained via DFT with experimental melting reaction data to train a machine learning model on 4,536 binary compounds.
Among the various machine learning algorithms tested, the XGBoost-based classification model demonstrated the highest accuracy in predicting whether alloys would mix well, achieving a prediction accuracy of approximately 82.5%. The team also applied the Shapley value method* to analyze the key features of the model. One major finding was that sharp changes in the slope of the formation energy curve (referred to as “convex hull sharpness”) were the most significant factor. A steep slope indicates a composition with energetically favorable (i.e., stable) formation.
*Shapley value: An explainability method in AI used to determine how much each feature contributed to a prediction.
The most notable significance of this study is that it predicts alloy melting behavior without performing high-temperature experiments. This is especially useful for materials such as high-entropy alloys or ultra-heat-resistant alloys, which are difficult to handle experimentally. The approach could also be extended to the design of complex multi-component alloy systems in the future.
Furthermore, the physical indicators identified by the AI model showed high consistency with actual experimental results on how well alloys mix and remain stable. This suggests that the model could be broadly applied to the development of various metal materials and the prediction of structural stability.
Professor Seungbum Hong of KAIST stated, “This research demonstrates how data-driven predictive materials development is possible by integrating computational methods, experimental data, and machine learning—departing from the traditional experience-based alloy design.” He added, “In the future, by incorporating state-of-the-art AI techniques such as generative models and reinforcement learning, we could enter an era where completely new alloys are designed automatically.”
<Model performance and feature importance analysis for predicting melting congruency. (a) SHAP summary plot showing the impact of individual features on model predictions. (b) Confusion matrix illustrating the model’s classification performance. (c) Receiver operating characteristic (ROC) curve with an AUC (area under the curve) score of 0.87, indicating a strong classification performance.>
Ph.D. candidate Youngwoo Choi, from the Department of Materials Science and Engineering at KAIST, participated as the first author. The study was published in the May issue of APL Machine Learning, a prestigious journal in the field of machine learning published by the American Institute of Physics, and was selected as a “Featured Article.”
※ Paper title: Machine learning-based melting congruency prediction of binary compounds using density functional theory-calculated formation energy ※ DOI: 10.1063/5.0247514
This research was supported by the Ministry of Science and ICT and the National Research Foundation of Korea.
2025.07.14 View 433 -
 Professor Jung-woo' Choi ‘s Team Comes in First at the World's Top Acoustic AI Challenge
<Photo1. (From left) Ph.D candidate Yong-hoo Kwon, M.S candidate Do-hwan Kim, Professor Jung-woo Choi, Dr. Dong-heon Lee>
'Acoustic separation and classification technology' is a next-generation artificial intelligence (AI) core technology that enables the early detection of abnormal sounds in areas such as drones, fault detection of factory pipelines, and border surveillance systems, or allows for the separation and editing of spatial audio by sound source when producing AR/VR content.
On the 11th of July, a research team led by Professor Jung-woo Choi of KAIST's Department of Electrical and Electronic Engineering won first place in the 'Spatial Semantic Segmentation of Sound Scenes' task of the 'DCASE2025 Challenge,' the world's most prestigious acoustic detection and analysis competition.
This year’s challenge featured 86 teams competing across six tasks. In this competition, the KAIST research team achieved the best performance in their first-ever participation to Task 4. Professor Jung-woo Choi’s research team consisted of Dr. Dong-heon, Lee, Ph.D. candidate Young-hoo Kwon, and M.S. candidate Do-hwan Kim.
Task 4 titled 'Spatial Semantic Segmentation of Sound Scenes' is a highly demanding task requiring the analysis of spatial information in multi-channel audio signals with overlapping sound sources. The goal was to separate individual sounds and classify them into 18 predefined categories. The research team plans to present their technology at the DCASE workshop in Barcelona this October.
<Figure 1. Example of an acoustic scene with multiple mixed sounds>
Early this year, Dr. Dong-heon Lee developed a state-of-the-art sound source separation AI that combines Transformer and Mamba architectures. During the competition, centered around researcher Young-hoo Kwon, they completed a ‘chain-of-inference architecture' AI model that performs sound source separation and classification again, using the waveforms and types of the initially separated sound sources as clues. This AI model is inspired by human’s auditory scene analysis mechanism that isolates individual sounds by focusing on incomplete clues such as sound type, rhythm, or direction, when listening to complex sounds.
Through this, the team was the only participant to achieve double-digit performance (11 dB) in 'Class-Aware Signal-to-Distortion Ratio Improvement (CA-SDRi)*,' which is the measure for ranking how well the AI separated and classified sounds, proving their technical excellence.
Class-Aware Signal-to-Distortion Ratio Improvement (CA-SDRi): Measures how much clearer (less distorted) the desired sound is separated and classified compared to the original audio, in dB (decibels). A higher number indicates more accurate and cleaner sound separation.
Prof. Jung-woo Choi remarked, "The research team has showcased world-leading acoustic separation AI models for the past three years, and I am delighted that these results have been officially recognized." He added, "I am proud of every member of the research team for winning first place through focused research, despite the significant increase in difficulty and having only a few weeks for development."
<Figure 2. Time-frequency patterns of sound sources separated from a mixed source>
The IEEE DCASE Challenge 2025 was held online, with submissions accepted from April 1 to June 15 and results announced on June 30. Since its launch in 2013, the DCASE Challenge has served as a premier global platform of IEEE Signal Processing Society for showcasing cutting-edge AI models in acoustic signal processing.
This research was supported by the Mid-Career Researcher Support Project and STEAM Research Project of the National Research Foundation of Korea, funded by the Ministry of Education, Science and Technology, as well as support from the Future Defense Research Center, funded by the Defense Acquisition Program Administration and the Agency for Defense Development.
2025.07.13 View 452
Professor Jung-woo' Choi ‘s Team Comes in First at the World's Top Acoustic AI Challenge
<Photo1. (From left) Ph.D candidate Yong-hoo Kwon, M.S candidate Do-hwan Kim, Professor Jung-woo Choi, Dr. Dong-heon Lee>
'Acoustic separation and classification technology' is a next-generation artificial intelligence (AI) core technology that enables the early detection of abnormal sounds in areas such as drones, fault detection of factory pipelines, and border surveillance systems, or allows for the separation and editing of spatial audio by sound source when producing AR/VR content.
On the 11th of July, a research team led by Professor Jung-woo Choi of KAIST's Department of Electrical and Electronic Engineering won first place in the 'Spatial Semantic Segmentation of Sound Scenes' task of the 'DCASE2025 Challenge,' the world's most prestigious acoustic detection and analysis competition.
This year’s challenge featured 86 teams competing across six tasks. In this competition, the KAIST research team achieved the best performance in their first-ever participation to Task 4. Professor Jung-woo Choi’s research team consisted of Dr. Dong-heon, Lee, Ph.D. candidate Young-hoo Kwon, and M.S. candidate Do-hwan Kim.
Task 4 titled 'Spatial Semantic Segmentation of Sound Scenes' is a highly demanding task requiring the analysis of spatial information in multi-channel audio signals with overlapping sound sources. The goal was to separate individual sounds and classify them into 18 predefined categories. The research team plans to present their technology at the DCASE workshop in Barcelona this October.
<Figure 1. Example of an acoustic scene with multiple mixed sounds>
Early this year, Dr. Dong-heon Lee developed a state-of-the-art sound source separation AI that combines Transformer and Mamba architectures. During the competition, centered around researcher Young-hoo Kwon, they completed a ‘chain-of-inference architecture' AI model that performs sound source separation and classification again, using the waveforms and types of the initially separated sound sources as clues. This AI model is inspired by human’s auditory scene analysis mechanism that isolates individual sounds by focusing on incomplete clues such as sound type, rhythm, or direction, when listening to complex sounds.
Through this, the team was the only participant to achieve double-digit performance (11 dB) in 'Class-Aware Signal-to-Distortion Ratio Improvement (CA-SDRi)*,' which is the measure for ranking how well the AI separated and classified sounds, proving their technical excellence.
Class-Aware Signal-to-Distortion Ratio Improvement (CA-SDRi): Measures how much clearer (less distorted) the desired sound is separated and classified compared to the original audio, in dB (decibels). A higher number indicates more accurate and cleaner sound separation.
Prof. Jung-woo Choi remarked, "The research team has showcased world-leading acoustic separation AI models for the past three years, and I am delighted that these results have been officially recognized." He added, "I am proud of every member of the research team for winning first place through focused research, despite the significant increase in difficulty and having only a few weeks for development."
<Figure 2. Time-frequency patterns of sound sources separated from a mixed source>
The IEEE DCASE Challenge 2025 was held online, with submissions accepted from April 1 to June 15 and results announced on June 30. Since its launch in 2013, the DCASE Challenge has served as a premier global platform of IEEE Signal Processing Society for showcasing cutting-edge AI models in acoustic signal processing.
This research was supported by the Mid-Career Researcher Support Project and STEAM Research Project of the National Research Foundation of Korea, funded by the Ministry of Education, Science and Technology, as well as support from the Future Defense Research Center, funded by the Defense Acquisition Program Administration and the Agency for Defense Development.
2025.07.13 View 452 -
 KAIST Kicks Off the Expansion of its Creative Learning Building, a 50th Anniversary Donation Landmark
KAIST announced on July 10th that it held a groundbreaking ceremony on July 9th for the expansion of its Creative Learning Building. This project, which celebrates the university's 50th anniversary, will become a significant donation-funded landmark and marks the official start of its construction.
<(From left) President Kwang Hyung Lee, Former President Sung-Chul Shin>
The groundbreaking ceremony was attended by key donors who graced the occasion, including KAIST President Kwang Hyung Lee, former President Sung-Chul Shin, Alumni Association President Yoon-Tae Lee, as well as parents and faculty member.
The Creative Learning Building serves as a primary space where KAIST undergraduate and graduate students attend lectures, functioning as a central hub for a variety of classes and talks. It also houses student support departments, including the Student Affairs Office, establishing itself as a student-centric complex that integrates educational, counseling, and welfare functions.
This expansion is more than just an increase in educational facilities; it's being developed as a "donation landmark" embodying KAIST's identity and future vision. Designed with a focus on creative convergence education, this project aims to create a new educational hub that organically combines education, exchange, and welfare functions
The campaign included over 230 participants, including KAIST alumni Byung-gyu Chang, Chairman of Krafton, former Alumni Association President Ki-chul Cha, Dr. Kun-mo Chung (former Minister of Science and Technology), as well as faculty members, parents, and current students. They collectively raised 6.5 billion KRW in donations. The total cost for this expansion project is 9 billion KRW, encompassing a gross floor area of 3,222.92㎡ across five above-ground floors, with completion targeted for September 2026.
2025.07.10 View 511
KAIST Kicks Off the Expansion of its Creative Learning Building, a 50th Anniversary Donation Landmark
KAIST announced on July 10th that it held a groundbreaking ceremony on July 9th for the expansion of its Creative Learning Building. This project, which celebrates the university's 50th anniversary, will become a significant donation-funded landmark and marks the official start of its construction.
<(From left) President Kwang Hyung Lee, Former President Sung-Chul Shin>
The groundbreaking ceremony was attended by key donors who graced the occasion, including KAIST President Kwang Hyung Lee, former President Sung-Chul Shin, Alumni Association President Yoon-Tae Lee, as well as parents and faculty member.
The Creative Learning Building serves as a primary space where KAIST undergraduate and graduate students attend lectures, functioning as a central hub for a variety of classes and talks. It also houses student support departments, including the Student Affairs Office, establishing itself as a student-centric complex that integrates educational, counseling, and welfare functions.
This expansion is more than just an increase in educational facilities; it's being developed as a "donation landmark" embodying KAIST's identity and future vision. Designed with a focus on creative convergence education, this project aims to create a new educational hub that organically combines education, exchange, and welfare functions
The campaign included over 230 participants, including KAIST alumni Byung-gyu Chang, Chairman of Krafton, former Alumni Association President Ki-chul Cha, Dr. Kun-mo Chung (former Minister of Science and Technology), as well as faculty members, parents, and current students. They collectively raised 6.5 billion KRW in donations. The total cost for this expansion project is 9 billion KRW, encompassing a gross floor area of 3,222.92㎡ across five above-ground floors, with completion targeted for September 2026.
2025.07.10 View 511