inhibition
-
 “One Experiment Is All It Takes”: KAIST Team Revolutionizes Drug Interaction Testing, Replacing 60,000 Studies
A groundbreaking new method developed by researchers at KAIST and Chungnam National University could drastically streamline drug interaction testing — replacing dozens of traditional experiments with just one.
The research, led by Professor Jae Kyoung Kim of KAIST Department of Mathematical Sciences & IBS Biomedical Mathematics Group and Professor Sang Kyum Kim of Chungnam National University's College of Pharmacy, introduces a novel analysis technique called 50-BOA, published in Nature Communications on June 5, 2025.
< Photo 1. (From left) Professor Sang Kyum Kim (Chungnam National University College of Pharmacy, co-corresponding author), Dr. Yun Min Song (IBS Biomedical Mathematics Group, formerly KAIST Department of Mathematical Sciences, co-first author), undergraduate student Hyeong Jun Jang (KAIST, co-first author), Professor Jae Kyoung Kim (KAIST and IBS Biomedical Mathematics Group, co-corresponding author) (Top left in the bubble) Professor Hwi-yeol Yun (Chungnam National University College of Pharmacy, co-author) >
For decades, scientists have had to repeat drug inhibition experiments across a wide range of concentrations to estimate inhibition constants — a process seen in over 60,000 scientific publications. But the KAIST-led team discovered that a single, well-chosen inhibitor concentration can yield even more accurate results.
< Figure 1. Graphical summary of 50-BOA. 50-BOA improves the accuracy and efficiency of inhibition constant estimation by using only a single inhibitor concentration instead of the traditionally used method of employing multiple inhibitor concentrations. >
“This approach challenges long-standing assumptions in experimental pharmacology,” says Prof. Kim. “It shows how mathematics can fundamentally redesign life science experiments.”
By mathematically analyzing the sources of error in conventional methods, the team found that over half the data typically collected adds no value or even skews results. Their new method not only cuts experimental effort by over 75%, but also enhances reproducibility and accuracy.
To help researchers adopt the method quickly, the team developed a user-friendly tool that takes simple Excel files as input, now freely available on GitHub:
☞ https://github.com/Mathbiomed/50-BOA
< Figure 2. The MATLAB and R package of 50-BOA at GitHub >
The work holds promise for faster and more reliable drug development, especially in assessing potential interactions in combination therapies. The U.S. FDA already emphasizes the importance of accurate enzyme inhibition assessment during early-stage drug evaluation — and this method could soon become a new gold standard.
2025.06.16 View 903
“One Experiment Is All It Takes”: KAIST Team Revolutionizes Drug Interaction Testing, Replacing 60,000 Studies
A groundbreaking new method developed by researchers at KAIST and Chungnam National University could drastically streamline drug interaction testing — replacing dozens of traditional experiments with just one.
The research, led by Professor Jae Kyoung Kim of KAIST Department of Mathematical Sciences & IBS Biomedical Mathematics Group and Professor Sang Kyum Kim of Chungnam National University's College of Pharmacy, introduces a novel analysis technique called 50-BOA, published in Nature Communications on June 5, 2025.
< Photo 1. (From left) Professor Sang Kyum Kim (Chungnam National University College of Pharmacy, co-corresponding author), Dr. Yun Min Song (IBS Biomedical Mathematics Group, formerly KAIST Department of Mathematical Sciences, co-first author), undergraduate student Hyeong Jun Jang (KAIST, co-first author), Professor Jae Kyoung Kim (KAIST and IBS Biomedical Mathematics Group, co-corresponding author) (Top left in the bubble) Professor Hwi-yeol Yun (Chungnam National University College of Pharmacy, co-author) >
For decades, scientists have had to repeat drug inhibition experiments across a wide range of concentrations to estimate inhibition constants — a process seen in over 60,000 scientific publications. But the KAIST-led team discovered that a single, well-chosen inhibitor concentration can yield even more accurate results.
< Figure 1. Graphical summary of 50-BOA. 50-BOA improves the accuracy and efficiency of inhibition constant estimation by using only a single inhibitor concentration instead of the traditionally used method of employing multiple inhibitor concentrations. >
“This approach challenges long-standing assumptions in experimental pharmacology,” says Prof. Kim. “It shows how mathematics can fundamentally redesign life science experiments.”
By mathematically analyzing the sources of error in conventional methods, the team found that over half the data typically collected adds no value or even skews results. Their new method not only cuts experimental effort by over 75%, but also enhances reproducibility and accuracy.
To help researchers adopt the method quickly, the team developed a user-friendly tool that takes simple Excel files as input, now freely available on GitHub:
☞ https://github.com/Mathbiomed/50-BOA
< Figure 2. The MATLAB and R package of 50-BOA at GitHub >
The work holds promise for faster and more reliable drug development, especially in assessing potential interactions in combination therapies. The U.S. FDA already emphasizes the importance of accurate enzyme inhibition assessment during early-stage drug evaluation — and this method could soon become a new gold standard.
2025.06.16 View 903 -
 KAIST Identifies Master Regulator Blocking Immunotherapy, Paving the Way for a New Lung Cancer Treatment
Immune checkpoint inhibitors, a class of immunotherapies that help immune cells attack cancer more effectively, have revolutionized cancer treatment. However, fewer than 20% of patients respond to these treatments, highlighting the urgent need for new strategies tailored to both responders and non-responders.
KAIST researchers have discovered that 'DEAD-box helicases 54 (DDX54)', a type of RNA-binding protein, is the master regulator that hinders the effectiveness of immunotherapy—opening a new path for lung cancer treatment. This breakthrough technology has been transferred to faculty startup BioRevert Inc., where it is currently being developed as a companion therapeutic and is expected to enter clinical trials by 2028.
< Photo 1. (From left) Researcher Jungeun Lee, Professor Kwang-Hyun Cho and Postdoctoral Researcher Jeong-Ryeol Gong of the Department of Bio and Brain Engineering at KAIST >
KAIST (represented by President Kwang-Hyung Lee) announced on April 8 that a research team led by Professor Kwang-Hyun Cho from the Department of Bio and Brain Engineering had identified DDX54 as a critical factor that determines the immune evasion capacity of lung cancer cells. They demonstrated that suppressing DDX54 enhances immune cell infiltration into tumors and significantly improves the efficacy of immunotherapy.
Immunotherapy using anti-PD-1 or anti-PD-L1 antibodies is considered a powerful approach in cancer treatment. However, its low response rate limits the number of patients who actually benefit.
To identify likely responders, tumor mutational burden (TMB) has recently been approved by the FDA as a key biomarker for immunotherapy. Cancers with high mutation rates are thought to be more responsive to immune checkpoint inhibitors. However, even tumors with high TMB can display an “immune-desert” phenotype—where immune cell infiltration is severely limited—resulting in poor treatment responses.
< Figure 1. DDX54 was identified as the master regulator that induces resistance to immunotherapy by orchestrating suppression of immune cell infiltration through cancer tissues as lung cancer cells become immune-evasive >
Professor Kwang-Hyun Cho's research team compared transcriptome and genome data of lung cancer patients with immune evasion capabilities through gene regulatory network analysis (A) and discovered DDX54, a master regulator that induces resistance to immunotherapy (B-F).
This study is especially significant in that it successfully demonstrated that suppressing DDX54 in immune-desert lung tumors can overcome immunotherapy resistance and improve treatment outcomes.
The team used transcriptomic and genomic data from immune-evasive lung cancer patients and employed systems biology techniques to infer gene regulatory networks. Through this analysis, they identified DDX54 as a central regulator in the immune evasion of lung cancer cells.
In a syngeneic mouse model, the suppression of DDX54 led to significant increases in the infiltration of anti-cancer immune cells such as T cells and NK cells, and greatly improved the response to immunotherapy.
Single-cell transcriptomic and spatial transcriptomic analyses further showed that combination therapy targeting DDX54 promoted the differentiation of T cells and memory T cells that suppress tumors, while reducing the infiltration of regulatory T cells and exhausted T cells that support tumor growth.
< Figure 2. In the syngeneic mouse model made of lung cancer cells, it was confirmed that inhibiting DDX54 reversed the immune-evasion ability of cancer cells and enhanced the sensitivity to anti-PD-1 therapy >
In a syngeneic mouse model made of lung cancer cells exhibiting immunotherapy resistance, the treatment applied after DDX54 inhibition resulted in statistically significant inhibition of lung cancer growth (B-D) and a significant increase in immune cell infiltration into the tumor tissue (E, F).
The mechanism is believed to involve DDX54 suppression inactivating signaling pathways such as JAK-STAT, MYC, and NF-κB, thereby downregulating immune-evasive proteins CD38 and CD47. This also reduced the infiltration of circulating monocytes—which promote tumor development—and promoted the differentiation of M1 macrophages that play anti-tumor roles.
Professor Kwang-Hyun Cho stated, “We have, for the first time, identified a master regulatory factor that enables immune evasion in lung cancer cells. By targeting this factor, we developed a new therapeutic strategy that can induce responsiveness to immunotherapy in previously resistant cancers.”
He added, “The discovery of DDX54—hidden within the complex molecular networks of cancer cells—was made possible through the systematic integration of systems biology, combining IT and BT.”
The study, led by Professor Kwang-Hyun Cho, was published in the Proceedings of the National Academy of Sciences of the United States of America (PNAS) on April 2, 2025, with Jeong-Ryeol Gong being the first author, Jungeun Lee, a co-first author, and Younghyun Han, a co-author of the article.
< Figure 3. Single-cell transcriptome and spatial transcriptome analysis confirmed that knockdown of DDX54 increased immune cell infiltration into cancer tissues >
In a syngeneic mouse model made of lung cancer cells that underwent immunotherapy in combination with DDX54 inhibition, single-cell transcriptome (H-L) and spatial transcriptome (A-G) analysis of immune cells infiltrating inside cancer tissues were performed. As a result, it was confirmed that anticancer immune cells such as T cells, B cells, and NK cells actively infiltrated the core of lung cancer tissues when DDX54 inhibition and immunotherapy were concurrently administered.
(Paper title: “DDX54 downregulation enhances anti-PD1 therapy in immune-desert lung tumors with high tumor mutational burden,” DOI: https://doi.org/10.1073/pnas.2412310122)
This work was supported by the Ministry of Science and ICT and the National Research Foundation of Korea through the Mid-Career Research Program and Basic Research Laboratory Program.
< Figure 4. The identified master regulator DDX54 was confirmed to induce CD38 and CD47 expression through Jak-Stat3, MYC, and NF-κB activation. >
DDX54 activates the Jak-Stat3, MYC, and NF-κB pathways in lung cancer cells to increase CD38 and CD47 expression (A-G). This creates a cancer microenvironment that contributes to cancer development (H) and ultimately induces immune anticancer treatment resistance.
< Figure 5. It was confirmed that an immune-inflamed environment can be created by combining DDX54 inhibition and immune checkpoint inhibitor (ICI) therapy. >
When DDX54 inhibition and ICI therapy are simultaneously administered, the cancer cell characteristics change, the immune evasion ability is restored, and the environment is transformed into an ‘immune-activated’ environment in which immune cells easily infiltrate cancer tissues. This strengthens the anticancer immune response, thereby increasing the sensitivity of immunotherapy even in lung cancer tissues that previously had low responsiveness to immunotherapy.
2025.04.08 View 4644
KAIST Identifies Master Regulator Blocking Immunotherapy, Paving the Way for a New Lung Cancer Treatment
Immune checkpoint inhibitors, a class of immunotherapies that help immune cells attack cancer more effectively, have revolutionized cancer treatment. However, fewer than 20% of patients respond to these treatments, highlighting the urgent need for new strategies tailored to both responders and non-responders.
KAIST researchers have discovered that 'DEAD-box helicases 54 (DDX54)', a type of RNA-binding protein, is the master regulator that hinders the effectiveness of immunotherapy—opening a new path for lung cancer treatment. This breakthrough technology has been transferred to faculty startup BioRevert Inc., where it is currently being developed as a companion therapeutic and is expected to enter clinical trials by 2028.
< Photo 1. (From left) Researcher Jungeun Lee, Professor Kwang-Hyun Cho and Postdoctoral Researcher Jeong-Ryeol Gong of the Department of Bio and Brain Engineering at KAIST >
KAIST (represented by President Kwang-Hyung Lee) announced on April 8 that a research team led by Professor Kwang-Hyun Cho from the Department of Bio and Brain Engineering had identified DDX54 as a critical factor that determines the immune evasion capacity of lung cancer cells. They demonstrated that suppressing DDX54 enhances immune cell infiltration into tumors and significantly improves the efficacy of immunotherapy.
Immunotherapy using anti-PD-1 or anti-PD-L1 antibodies is considered a powerful approach in cancer treatment. However, its low response rate limits the number of patients who actually benefit.
To identify likely responders, tumor mutational burden (TMB) has recently been approved by the FDA as a key biomarker for immunotherapy. Cancers with high mutation rates are thought to be more responsive to immune checkpoint inhibitors. However, even tumors with high TMB can display an “immune-desert” phenotype—where immune cell infiltration is severely limited—resulting in poor treatment responses.
< Figure 1. DDX54 was identified as the master regulator that induces resistance to immunotherapy by orchestrating suppression of immune cell infiltration through cancer tissues as lung cancer cells become immune-evasive >
Professor Kwang-Hyun Cho's research team compared transcriptome and genome data of lung cancer patients with immune evasion capabilities through gene regulatory network analysis (A) and discovered DDX54, a master regulator that induces resistance to immunotherapy (B-F).
This study is especially significant in that it successfully demonstrated that suppressing DDX54 in immune-desert lung tumors can overcome immunotherapy resistance and improve treatment outcomes.
The team used transcriptomic and genomic data from immune-evasive lung cancer patients and employed systems biology techniques to infer gene regulatory networks. Through this analysis, they identified DDX54 as a central regulator in the immune evasion of lung cancer cells.
In a syngeneic mouse model, the suppression of DDX54 led to significant increases in the infiltration of anti-cancer immune cells such as T cells and NK cells, and greatly improved the response to immunotherapy.
Single-cell transcriptomic and spatial transcriptomic analyses further showed that combination therapy targeting DDX54 promoted the differentiation of T cells and memory T cells that suppress tumors, while reducing the infiltration of regulatory T cells and exhausted T cells that support tumor growth.
< Figure 2. In the syngeneic mouse model made of lung cancer cells, it was confirmed that inhibiting DDX54 reversed the immune-evasion ability of cancer cells and enhanced the sensitivity to anti-PD-1 therapy >
In a syngeneic mouse model made of lung cancer cells exhibiting immunotherapy resistance, the treatment applied after DDX54 inhibition resulted in statistically significant inhibition of lung cancer growth (B-D) and a significant increase in immune cell infiltration into the tumor tissue (E, F).
The mechanism is believed to involve DDX54 suppression inactivating signaling pathways such as JAK-STAT, MYC, and NF-κB, thereby downregulating immune-evasive proteins CD38 and CD47. This also reduced the infiltration of circulating monocytes—which promote tumor development—and promoted the differentiation of M1 macrophages that play anti-tumor roles.
Professor Kwang-Hyun Cho stated, “We have, for the first time, identified a master regulatory factor that enables immune evasion in lung cancer cells. By targeting this factor, we developed a new therapeutic strategy that can induce responsiveness to immunotherapy in previously resistant cancers.”
He added, “The discovery of DDX54—hidden within the complex molecular networks of cancer cells—was made possible through the systematic integration of systems biology, combining IT and BT.”
The study, led by Professor Kwang-Hyun Cho, was published in the Proceedings of the National Academy of Sciences of the United States of America (PNAS) on April 2, 2025, with Jeong-Ryeol Gong being the first author, Jungeun Lee, a co-first author, and Younghyun Han, a co-author of the article.
< Figure 3. Single-cell transcriptome and spatial transcriptome analysis confirmed that knockdown of DDX54 increased immune cell infiltration into cancer tissues >
In a syngeneic mouse model made of lung cancer cells that underwent immunotherapy in combination with DDX54 inhibition, single-cell transcriptome (H-L) and spatial transcriptome (A-G) analysis of immune cells infiltrating inside cancer tissues were performed. As a result, it was confirmed that anticancer immune cells such as T cells, B cells, and NK cells actively infiltrated the core of lung cancer tissues when DDX54 inhibition and immunotherapy were concurrently administered.
(Paper title: “DDX54 downregulation enhances anti-PD1 therapy in immune-desert lung tumors with high tumor mutational burden,” DOI: https://doi.org/10.1073/pnas.2412310122)
This work was supported by the Ministry of Science and ICT and the National Research Foundation of Korea through the Mid-Career Research Program and Basic Research Laboratory Program.
< Figure 4. The identified master regulator DDX54 was confirmed to induce CD38 and CD47 expression through Jak-Stat3, MYC, and NF-κB activation. >
DDX54 activates the Jak-Stat3, MYC, and NF-κB pathways in lung cancer cells to increase CD38 and CD47 expression (A-G). This creates a cancer microenvironment that contributes to cancer development (H) and ultimately induces immune anticancer treatment resistance.
< Figure 5. It was confirmed that an immune-inflamed environment can be created by combining DDX54 inhibition and immune checkpoint inhibitor (ICI) therapy. >
When DDX54 inhibition and ICI therapy are simultaneously administered, the cancer cell characteristics change, the immune evasion ability is restored, and the environment is transformed into an ‘immune-activated’ environment in which immune cells easily infiltrate cancer tissues. This strengthens the anticancer immune response, thereby increasing the sensitivity of immunotherapy even in lung cancer tissues that previously had low responsiveness to immunotherapy.
2025.04.08 View 4644 -
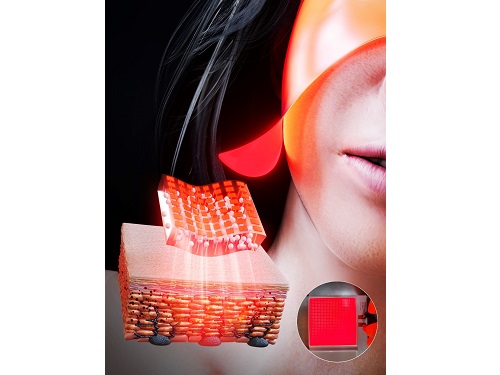 KAIST Team Develops Surface-Lighting MicroLED Patch with Significant Melanogenesis Inhibition Effect
A KAIST research team led by Ph.d candidate Jae Hee Lee and Professor Keon Jae Lee from the Department of Materials Science and Engineering has developed a surface-lighting microLED patch for UV-induced melanogenesis inhibition.
Melanin is brown or dark pigments existing in the skin, which can be abnormally synthesized by external UV or stress. Since the excessive melanin leads to skin diseases such as spots and freckles, proper treatment is required to return normal skin condition.
Recently, LED-based photo-stimulators have been released for skin care, however, their therapeutic effect is still controversial. Since conventional LED stimulators cannot conformally attach to the human skin, distance-induced side effects are caused by light loss and high heat transfer. To achieve effective phototreatment, the LED stimulator needs to be irradiated in contact with the human skin surface, enabling proper and uniform light deliver to the dermis with minimal optical loss.
In this work, the research team fabricated skin-attachable surface-lighting microLED (SµLED, 4 × 4 cm2) patch by utilizing a thousand of microLED chips and silica-embedded light diffusion layer. 100 µm-sized LED chips are vertically-interconnected for high flexibility and low heat generation, allowing its long-term operation on the human skin.
< Image 1. The overall concept of SµLED patch. a) SµLED patch operated on the human skin. b) Schematic illustration of SµLED patch structure. c) 4 × 4 cm2-sized SµLED patch. d) Schematic illustration of the advantages of SµLED patch such as efficient light delivery, low heat generation, and surface-lighting irradiation. >
The research team confirmed melanogenesis inhibition by irradiating the SµLED patch and the conventional LED (CLED) on the artificial human skin and mice dorsal skin. The SµLED-treated groups of human cells and mouse tissues showed minimal epidermal photo-toxicity and consistently effective reduction in synthesized melanin, compared to CLED-treated groups. In addition, significant suppression of proteins/catalysts expression involved in melanin synthesis such as MITF (microphthalmia-associated transcription factor), Melan-A and tyrosinase was verified.
< Image 2. The efficacy of melanogenesis inhibition on 3D human skin cells. a). Different irradiation conditions for a-MSH (major factor to stimulate melanin synthesis) treated cells. b) The ratio of pigmented area to total epidermis area. c) Relative variance of melanin level in 1 cm2-sized skin cells. A low variance means that melanin is evenly distributed, and a high variance means that the melanin is irregularly distributed. d) Optical images after in vitro experiments for 12 days. Scale bar, 1cm. e) Histological analysis of 3D skin, showing the greatest reduction in melanin after SµLED irradiation. Scale bar, 20 µm. >
< Image 3. The efficacy of melanogenesis inhibition on mouse dorsal skin. a) Optical images of mice dorsal skin after photo-treatment for 20 days. b) Histological analysis of mice dorsal skin. Less brown color means less expression of protein/catalysis involved in melanin synthesis. Scale bar, 50 µm. >
Prof. Keon Jae Lee said, “Our inorganic-based SµLED patch has outstanding characteristics in light efficiency, reliability, and durability. The SµLED patch is expected to give a great impact on the cosmetic field by reducing side effects and maximizing phototherapeutic effects.” The core technology of cosmetic SµLED has been transferred to Fronics co., Ltd, founded by Prof. Lee. Fronics is building foundry and equipment for mass production of SµLED masks for whole face cover and plans to release the products in March next year.
This paper entitled “Wearable Surface-Lighting Micro-Light-Emitting Diode Patch for Melanogenesis Inhibition” was published in the November 2022 issue of Advanced Healthcare Materials.
2022.11.22 View 12797
KAIST Team Develops Surface-Lighting MicroLED Patch with Significant Melanogenesis Inhibition Effect
A KAIST research team led by Ph.d candidate Jae Hee Lee and Professor Keon Jae Lee from the Department of Materials Science and Engineering has developed a surface-lighting microLED patch for UV-induced melanogenesis inhibition.
Melanin is brown or dark pigments existing in the skin, which can be abnormally synthesized by external UV or stress. Since the excessive melanin leads to skin diseases such as spots and freckles, proper treatment is required to return normal skin condition.
Recently, LED-based photo-stimulators have been released for skin care, however, their therapeutic effect is still controversial. Since conventional LED stimulators cannot conformally attach to the human skin, distance-induced side effects are caused by light loss and high heat transfer. To achieve effective phototreatment, the LED stimulator needs to be irradiated in contact with the human skin surface, enabling proper and uniform light deliver to the dermis with minimal optical loss.
In this work, the research team fabricated skin-attachable surface-lighting microLED (SµLED, 4 × 4 cm2) patch by utilizing a thousand of microLED chips and silica-embedded light diffusion layer. 100 µm-sized LED chips are vertically-interconnected for high flexibility and low heat generation, allowing its long-term operation on the human skin.
< Image 1. The overall concept of SµLED patch. a) SµLED patch operated on the human skin. b) Schematic illustration of SµLED patch structure. c) 4 × 4 cm2-sized SµLED patch. d) Schematic illustration of the advantages of SµLED patch such as efficient light delivery, low heat generation, and surface-lighting irradiation. >
The research team confirmed melanogenesis inhibition by irradiating the SµLED patch and the conventional LED (CLED) on the artificial human skin and mice dorsal skin. The SµLED-treated groups of human cells and mouse tissues showed minimal epidermal photo-toxicity and consistently effective reduction in synthesized melanin, compared to CLED-treated groups. In addition, significant suppression of proteins/catalysts expression involved in melanin synthesis such as MITF (microphthalmia-associated transcription factor), Melan-A and tyrosinase was verified.
< Image 2. The efficacy of melanogenesis inhibition on 3D human skin cells. a). Different irradiation conditions for a-MSH (major factor to stimulate melanin synthesis) treated cells. b) The ratio of pigmented area to total epidermis area. c) Relative variance of melanin level in 1 cm2-sized skin cells. A low variance means that melanin is evenly distributed, and a high variance means that the melanin is irregularly distributed. d) Optical images after in vitro experiments for 12 days. Scale bar, 1cm. e) Histological analysis of 3D skin, showing the greatest reduction in melanin after SµLED irradiation. Scale bar, 20 µm. >
< Image 3. The efficacy of melanogenesis inhibition on mouse dorsal skin. a) Optical images of mice dorsal skin after photo-treatment for 20 days. b) Histological analysis of mice dorsal skin. Less brown color means less expression of protein/catalysis involved in melanin synthesis. Scale bar, 50 µm. >
Prof. Keon Jae Lee said, “Our inorganic-based SµLED patch has outstanding characteristics in light efficiency, reliability, and durability. The SµLED patch is expected to give a great impact on the cosmetic field by reducing side effects and maximizing phototherapeutic effects.” The core technology of cosmetic SµLED has been transferred to Fronics co., Ltd, founded by Prof. Lee. Fronics is building foundry and equipment for mass production of SµLED masks for whole face cover and plans to release the products in March next year.
This paper entitled “Wearable Surface-Lighting Micro-Light-Emitting Diode Patch for Melanogenesis Inhibition” was published in the November 2022 issue of Advanced Healthcare Materials.
2022.11.22 View 12797