Department+of+Chemical
-
 An efficient strategy for developing microbial cell factories by employing synthetic small regulatory RNAs
A new metabolic engineering tool that allows fine control of gene expression level by employing synthetic small regulatory RNAs was developed to efficiently construct microbial cell factories producing desired chemicals and materials
Biotechnologists have been working hard to address the climate change and limited fossil resource issues through the development of sustainable processes for the production of chemicals, fuels and materials from renewable non-food biomass. One promising sustainable technology is the use of microbial cell factories for the efficient production of desired chemicals and materials. When microorganisms are isolated from nature, the performance in producing our desired product is rather poor. That is why metabolic engineering is performed to improve the metabolic and cellular characteristics to achieve enhanced production of desired product at high yield and productivity. Since the performance of microbial cell factory is very important in lowering the overall production cost of the bioprocess, many different strategies and tools have been developed for the metabolic engineering of microorganisms.
One of the big challenges in metabolic engineering is to find the best platform organism and to find those genes to be engineered so as to maximize the production efficiency of the desired chemical. Even Escherichia coli, the most widely utilized simple microorganism, has thousands of genes, the expression of which is highly regulated and interconnected to finely control cellular and metabolic activities. Thus, the complexity of cellular genetic interactions is beyond our intuition and thus it is very difficult to find effective target genes to engineer. Together with gene amplification strategy, gene knockout strategy has been an essential tool in metabolic engineering to redirect the pathway fluxes toward our desired product formation. However, experiment to engineer many genes can be rather difficult due to the time and effort required; for example, gene deletion experiment can take a few weeks depending on the microorganisms. Furthermore, as certain genes are essential or play important roles for the survival of a microorganism, gene knockout experiments cannot be performed. Even worse, there are many different microbial strains one can employ. There are more than 50 different E. coli strains that metabolic engineer can consider to use. Since gene knockout experiment is hard-coded (that is, one should repeat the gene knockout experiments for each strain), the result cannot be easily transferred from one strain to another.
A paper published in Nature Biotechnology online today addresses this issue and suggests a new strategy for identifying gene targets to be knocked out or knocked down through the use of synthetic small RNA. A Korean research team led by Distinguished Professor Sang Yup Lee at the Department of Chemical and Biomolecular Engineering, Korea Advanced Institute of Science and Technology (KAIST), a prestigeous science and engineering university in Korea reported that synthetic small RNA can be employed for finely controlling the expression levels of multiple genes at the translation level. Already well-known for their systems metabolic engineering strategies, Professor Lee’s team added one more strategy to efficiently develop microbial cell factories for the production of chemicals and materials.
Gene expression works like this: the hard-coded blueprint (DNA) is transcribed into messenger RNA (mRNA), and the coding information in mRNA is read to produce protein by ribosomes. Conventional genetic engineering approaches have often targeted modification of the blueprint itself (DNA) to alter organism’s physiological characteristics. Again, engineering the blueprint itself takes much time and effort, and in addition, the results obtained cannot be transferred to another organism without repeating the whole set of experiments. This is why Professor Lee and his colleagues aimed at controlling the gene expression level at the translation stage through the use of synthetic small RNA. They created novel RNAs that can regulate the translation of multiple messenger RNAs (mRNA), and consequently varying the expression levels of multiple genes at the same time. Briefly, synthetic regulatory RNAs interrupt gene expression process from DNA to protein by destroying the messenger RNAs to different yet controllable extents. The advantages of taking this strategy of employing synthetic small regulatory RNAs include simple, easy and high-throughput identification of gene knockout or knockdown targets, fine control of gene expression levels, transferability to many different host strains, and possibility of identifying those gene targets that are essential.
As proof-of-concept demonstration of the usefulness of this strategy, Professor Lee and his colleagues applied it to develop engineered E. coli strains capable of producing an aromatic amino acid tyrosine, which is used for stress symptom relief, food supplements, and precursor for many drugs. They examined a large number of genes in multiple E. coli strains, and developed a highly efficient tyrosine producer. Also, they were able to show that this strategy can be employed to an already metabolically engineered E. coli strain for further improvement by demonstrating the development of highly efficient producer of cadaverine, an important platform chemical for nylon in the chemical industry.
This new strategy, being simple yet very powerful for systems metabolic engineering, is thus expected to facilitate the efficient development of microbial cell factories capable of producing chemicals, fuels and materials from renewable biomass.
Source: Dokyun Na, Seung Min Yoo, Hannah Chung, Hyegwon Park, Jin Hwan Park, and Sang Yup Lee, “Metabolic engineering of Escherichia coli using synthetic small regulatory RNAs”, Nature Biotechnology, doi:10.1038/nbt.2461 (2013)
2013.03.19 View 11735
An efficient strategy for developing microbial cell factories by employing synthetic small regulatory RNAs
A new metabolic engineering tool that allows fine control of gene expression level by employing synthetic small regulatory RNAs was developed to efficiently construct microbial cell factories producing desired chemicals and materials
Biotechnologists have been working hard to address the climate change and limited fossil resource issues through the development of sustainable processes for the production of chemicals, fuels and materials from renewable non-food biomass. One promising sustainable technology is the use of microbial cell factories for the efficient production of desired chemicals and materials. When microorganisms are isolated from nature, the performance in producing our desired product is rather poor. That is why metabolic engineering is performed to improve the metabolic and cellular characteristics to achieve enhanced production of desired product at high yield and productivity. Since the performance of microbial cell factory is very important in lowering the overall production cost of the bioprocess, many different strategies and tools have been developed for the metabolic engineering of microorganisms.
One of the big challenges in metabolic engineering is to find the best platform organism and to find those genes to be engineered so as to maximize the production efficiency of the desired chemical. Even Escherichia coli, the most widely utilized simple microorganism, has thousands of genes, the expression of which is highly regulated and interconnected to finely control cellular and metabolic activities. Thus, the complexity of cellular genetic interactions is beyond our intuition and thus it is very difficult to find effective target genes to engineer. Together with gene amplification strategy, gene knockout strategy has been an essential tool in metabolic engineering to redirect the pathway fluxes toward our desired product formation. However, experiment to engineer many genes can be rather difficult due to the time and effort required; for example, gene deletion experiment can take a few weeks depending on the microorganisms. Furthermore, as certain genes are essential or play important roles for the survival of a microorganism, gene knockout experiments cannot be performed. Even worse, there are many different microbial strains one can employ. There are more than 50 different E. coli strains that metabolic engineer can consider to use. Since gene knockout experiment is hard-coded (that is, one should repeat the gene knockout experiments for each strain), the result cannot be easily transferred from one strain to another.
A paper published in Nature Biotechnology online today addresses this issue and suggests a new strategy for identifying gene targets to be knocked out or knocked down through the use of synthetic small RNA. A Korean research team led by Distinguished Professor Sang Yup Lee at the Department of Chemical and Biomolecular Engineering, Korea Advanced Institute of Science and Technology (KAIST), a prestigeous science and engineering university in Korea reported that synthetic small RNA can be employed for finely controlling the expression levels of multiple genes at the translation level. Already well-known for their systems metabolic engineering strategies, Professor Lee’s team added one more strategy to efficiently develop microbial cell factories for the production of chemicals and materials.
Gene expression works like this: the hard-coded blueprint (DNA) is transcribed into messenger RNA (mRNA), and the coding information in mRNA is read to produce protein by ribosomes. Conventional genetic engineering approaches have often targeted modification of the blueprint itself (DNA) to alter organism’s physiological characteristics. Again, engineering the blueprint itself takes much time and effort, and in addition, the results obtained cannot be transferred to another organism without repeating the whole set of experiments. This is why Professor Lee and his colleagues aimed at controlling the gene expression level at the translation stage through the use of synthetic small RNA. They created novel RNAs that can regulate the translation of multiple messenger RNAs (mRNA), and consequently varying the expression levels of multiple genes at the same time. Briefly, synthetic regulatory RNAs interrupt gene expression process from DNA to protein by destroying the messenger RNAs to different yet controllable extents. The advantages of taking this strategy of employing synthetic small regulatory RNAs include simple, easy and high-throughput identification of gene knockout or knockdown targets, fine control of gene expression levels, transferability to many different host strains, and possibility of identifying those gene targets that are essential.
As proof-of-concept demonstration of the usefulness of this strategy, Professor Lee and his colleagues applied it to develop engineered E. coli strains capable of producing an aromatic amino acid tyrosine, which is used for stress symptom relief, food supplements, and precursor for many drugs. They examined a large number of genes in multiple E. coli strains, and developed a highly efficient tyrosine producer. Also, they were able to show that this strategy can be employed to an already metabolically engineered E. coli strain for further improvement by demonstrating the development of highly efficient producer of cadaverine, an important platform chemical for nylon in the chemical industry.
This new strategy, being simple yet very powerful for systems metabolic engineering, is thus expected to facilitate the efficient development of microbial cell factories capable of producing chemicals, fuels and materials from renewable biomass.
Source: Dokyun Na, Seung Min Yoo, Hannah Chung, Hyegwon Park, Jin Hwan Park, and Sang Yup Lee, “Metabolic engineering of Escherichia coli using synthetic small regulatory RNAs”, Nature Biotechnology, doi:10.1038/nbt.2461 (2013)
2013.03.19 View 11735 -
 High Efficiency Bio-butanol production technology developed
KAIST and Korean Company cooperative research team has developed the technology that increases the productivity of bio-butanol to equal that of bio-ethanol and decreases the cost of production.
Professor Lee Sang Yeop (Department of Biological-Chemical Engineering) collaborated with GS Caltex and BioFuelChem Ltd. to develop a bio-butanol production process using the system metabolism engineering method that increased the productivity and decreased the production cost.
Bio-butanol is being widely regarded as the environmentally friendly next generation energy source that surpasses bio-ethanol.
The energy density of bio-butanol is 29.9MJ (mega Joule) per Liter, 48% larger than bio-ethanol (19.6MJ) and comparable to gasoline (32MJ). Bio-butanol is advantageous in that it can be processed from inedible biomass and is therefore unrelated to food crises.
Especially because bio-butanol shows similar characteristics especially in its octane rating, enthalpy of vaporization, and air-fuel ratio, it can be used in a gasoline engine.
However barriers such as difficulty in gene manipulation of producer bacterium and insufficient information prevented the mass production of bio-butanol.
Professor Lee’s team applied the system metabolism engineering method that he had invented to shift the focus to the production pathway of bio-butanol and made a new metabolism model.
In the new model the bio-butanol production pathway is divided into the hot channel and the cold channel.
The research team focused on improving the efficiency of the hot channel and succeeded in improving the product yield of 49% (compared to theoretical yield) to 87%.
The team furthered their research and developed a live bio-butanol collection and removal system with GS Caltex. The collaboration succeeded in producing 585g of butanol using 1.8kg of glucose at a rate of 1.3g per hour, boasting world’s highest concentration, productivity, and rate and improving productivity of fermentation by three fold and decreasing costs by 30%.
The result of the research was published in world renowned ‘mBio’ microbiology journal.
2012.12.21 View 10392
High Efficiency Bio-butanol production technology developed
KAIST and Korean Company cooperative research team has developed the technology that increases the productivity of bio-butanol to equal that of bio-ethanol and decreases the cost of production.
Professor Lee Sang Yeop (Department of Biological-Chemical Engineering) collaborated with GS Caltex and BioFuelChem Ltd. to develop a bio-butanol production process using the system metabolism engineering method that increased the productivity and decreased the production cost.
Bio-butanol is being widely regarded as the environmentally friendly next generation energy source that surpasses bio-ethanol.
The energy density of bio-butanol is 29.9MJ (mega Joule) per Liter, 48% larger than bio-ethanol (19.6MJ) and comparable to gasoline (32MJ). Bio-butanol is advantageous in that it can be processed from inedible biomass and is therefore unrelated to food crises.
Especially because bio-butanol shows similar characteristics especially in its octane rating, enthalpy of vaporization, and air-fuel ratio, it can be used in a gasoline engine.
However barriers such as difficulty in gene manipulation of producer bacterium and insufficient information prevented the mass production of bio-butanol.
Professor Lee’s team applied the system metabolism engineering method that he had invented to shift the focus to the production pathway of bio-butanol and made a new metabolism model.
In the new model the bio-butanol production pathway is divided into the hot channel and the cold channel.
The research team focused on improving the efficiency of the hot channel and succeeded in improving the product yield of 49% (compared to theoretical yield) to 87%.
The team furthered their research and developed a live bio-butanol collection and removal system with GS Caltex. The collaboration succeeded in producing 585g of butanol using 1.8kg of glucose at a rate of 1.3g per hour, boasting world’s highest concentration, productivity, and rate and improving productivity of fermentation by three fold and decreasing costs by 30%.
The result of the research was published in world renowned ‘mBio’ microbiology journal.
2012.12.21 View 10392 -
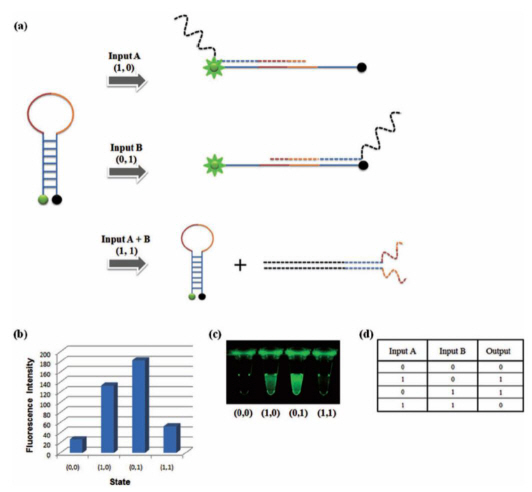 DNA based semiconductor technology developed
Professor Park Hyun Gyu’s research team from the Department of Chemical and Biomolecular Engineering at KAIST has successfully implemented all logic gates using DNA, a feat that led the research to be published as the cover paper for the international nanotechnology paper "Small".
Even with the latest technology, it was impossible to create a silicon based semiconductor smaller than 10nm, but because DNA has a thickness of only 2nm, this could lead to the creation of semiconductors with groundbreaking degrees of integration.
A 2 nm semiconductor will be able to store 10,000 HD movies within a size of a postage stamp, at least 100 times more than the current 20nm semiconductors.
DNAs are comprised of 4 bases which are continually connected: Adenine (A) with Thymine (T), and Guanine (G) with Cytosine (C).
For this research, the team used the specific binding properties of DNA, which forms its helix-shape, and a circular molecular beacon that has fluorescent signaling properties under structural changes.
The research team used input signals to open and close the circular DNA, the same principle that is applied to logic gates in digital circuits.
The output signal was measured using the increase and decrease of the fluorescent signal from the molecular beacon due to the opening and closing of the circular DNA respectively.
The team overcame the limited system problems of the existing logic gates and managed to implement all 8 logic gates (AND, OR, XOR, INHIBIT, NAND, NOR, XNOR, IMPlCATION). A multilevel circuit that connects different logic gates was also tested to show its regenerative properties.
Professor Park said that “cheap bio-electric devices with high degrees of integration will be made possible by this research” and that “there will be a large difference in the field of molecular level electronic research”
Mr. Park Gi Su, a doctoral candidate and the 1st author of this research, said that “a DNA sequence of 10 bases is only 3.4nm long and 2nm thick, which can be used to effectively increase the degree of integration of electronic devices” and that “a bio computer could materialize in the near future through DNA semiconductors with accurate logic gates”.
XOR Gate: The output signal 1 comes through the open circular DNA when either input DNA A or input DNA B is present. When both inputs are not present, the flourescent signal does not come through
2012.09.27 View 11387
DNA based semiconductor technology developed
Professor Park Hyun Gyu’s research team from the Department of Chemical and Biomolecular Engineering at KAIST has successfully implemented all logic gates using DNA, a feat that led the research to be published as the cover paper for the international nanotechnology paper "Small".
Even with the latest technology, it was impossible to create a silicon based semiconductor smaller than 10nm, but because DNA has a thickness of only 2nm, this could lead to the creation of semiconductors with groundbreaking degrees of integration.
A 2 nm semiconductor will be able to store 10,000 HD movies within a size of a postage stamp, at least 100 times more than the current 20nm semiconductors.
DNAs are comprised of 4 bases which are continually connected: Adenine (A) with Thymine (T), and Guanine (G) with Cytosine (C).
For this research, the team used the specific binding properties of DNA, which forms its helix-shape, and a circular molecular beacon that has fluorescent signaling properties under structural changes.
The research team used input signals to open and close the circular DNA, the same principle that is applied to logic gates in digital circuits.
The output signal was measured using the increase and decrease of the fluorescent signal from the molecular beacon due to the opening and closing of the circular DNA respectively.
The team overcame the limited system problems of the existing logic gates and managed to implement all 8 logic gates (AND, OR, XOR, INHIBIT, NAND, NOR, XNOR, IMPlCATION). A multilevel circuit that connects different logic gates was also tested to show its regenerative properties.
Professor Park said that “cheap bio-electric devices with high degrees of integration will be made possible by this research” and that “there will be a large difference in the field of molecular level electronic research”
Mr. Park Gi Su, a doctoral candidate and the 1st author of this research, said that “a DNA sequence of 10 bases is only 3.4nm long and 2nm thick, which can be used to effectively increase the degree of integration of electronic devices” and that “a bio computer could materialize in the near future through DNA semiconductors with accurate logic gates”.
XOR Gate: The output signal 1 comes through the open circular DNA when either input DNA A or input DNA B is present. When both inputs are not present, the flourescent signal does not come through
2012.09.27 View 11387 -
 Production of chemicals without petroleum
Systems metabolic engineering of microorganisms allows efficient production of natural and non-natural chemicals from renewable non-food biomass
In our everyday life, we use gasoline, diesel, plastics, rubbers, and numerous chemicals that are derived from fossil oil through petrochemical refinery processes. However, this is not sustainable due to the limited nature of fossil resources. Furthermore, our world is facing problems associated with climate change and other environmental problems due to the increasing use of fossil resources. One solution to address above problems is the use of renewable non-food biomass for the production of chemicals, fuels and materials through biorefineries. Microorganisms are used as biocatalysts for converting biomass to the products of interest. However, when microorganisms are isolated from nature, their efficiencies of producing our desired chemicals and materials are rather low. Metabolic engineering is thus performed to improve cellular characteristics to desired levels. Over the last decade, much advances have been made in systems biology that allows system-wide characterization of cellular networks, both qualitatively and quantitatively, followed by whole-cell level engineering based on these findings. Furthermore, rapid advances in synthetic biology allow design and synthesis of fine controlled metabolic and gene regulatory circuits. The strategies and methods of systems biology and synthetic biology are rapidly integrated with metabolic engineering, thus resulting in "systems metabolic engineering".
In the paper published online in Nature Chemical Biology on May 17, Professor Sang Yup Lee and his colleagues at the Department of Chemical and Biomolecular Engineering, Korea Advanced Institute of Science and Technology (KAIST), Daejeon, Korea present new general strategies of systems metabolic engineering for developing microorganisms for the production of natural and non-natural chemicals from renewable biomass. They first classified the chemicals to be produced into four categories based on whether they have thus far been identified to exist in nature (natural vs. nonnatural) and whether they can be produced by inherent pathways of microorganisms (inherent, noninherent, or created): natural-inherent, natural-noninherent, non-natural-noninherent, and non-natural-created ones. General strategies for systems metabolic engineering of microorganisms for the production of these chemicals using various tools and methods based on omics, genome-scale metabolic modeling and simulation, evolutionary engineering, synthetic biology are suggested with relevant examples. For the production of non-natural chemicals, strategies for the construction of synthetic metabolic pathways are also suggested. Having collected diverse tools and methods for systems metabolic engineering, authors also suggest how to use them and their possible limitations.
Professor Sang Yup Lee said "It is expected that increasing number of chemicals and materials will be produced through biorefineries. We are now equipped with new strategies for developing microbial strains that can produce our desired products at very high efficiencies, thus allowing cost competitiveness to those produced by petrochemical refineries."
Editor of Nature Chemical Biology, Dr. Catherine Goodman, said "It is exciting to see how quickly science is progressing in this field – ideas that used to be science fiction are taking shape in research labs and biorefineries. The article by Professor Lee and his colleagues not only highlights the most advanced techniques and strategies available, but offers critical advice to progress the field as a whole."
The works of Professor Lee have been supported by the Advanced Biomass Center and Intelligent Synthetic Biology Center of Global Frontier Program from the Korean Ministry of Education, Science and Technology through National Research Foundation.
Contact: Dr. Sang Yup Lee, Distinguished Professor and Dean, KAIST, Daejeon, Korea (leesy@kaist.ac.kr, +82-42-350-3930)
2012.05.23 View 16069
Production of chemicals without petroleum
Systems metabolic engineering of microorganisms allows efficient production of natural and non-natural chemicals from renewable non-food biomass
In our everyday life, we use gasoline, diesel, plastics, rubbers, and numerous chemicals that are derived from fossil oil through petrochemical refinery processes. However, this is not sustainable due to the limited nature of fossil resources. Furthermore, our world is facing problems associated with climate change and other environmental problems due to the increasing use of fossil resources. One solution to address above problems is the use of renewable non-food biomass for the production of chemicals, fuels and materials through biorefineries. Microorganisms are used as biocatalysts for converting biomass to the products of interest. However, when microorganisms are isolated from nature, their efficiencies of producing our desired chemicals and materials are rather low. Metabolic engineering is thus performed to improve cellular characteristics to desired levels. Over the last decade, much advances have been made in systems biology that allows system-wide characterization of cellular networks, both qualitatively and quantitatively, followed by whole-cell level engineering based on these findings. Furthermore, rapid advances in synthetic biology allow design and synthesis of fine controlled metabolic and gene regulatory circuits. The strategies and methods of systems biology and synthetic biology are rapidly integrated with metabolic engineering, thus resulting in "systems metabolic engineering".
In the paper published online in Nature Chemical Biology on May 17, Professor Sang Yup Lee and his colleagues at the Department of Chemical and Biomolecular Engineering, Korea Advanced Institute of Science and Technology (KAIST), Daejeon, Korea present new general strategies of systems metabolic engineering for developing microorganisms for the production of natural and non-natural chemicals from renewable biomass. They first classified the chemicals to be produced into four categories based on whether they have thus far been identified to exist in nature (natural vs. nonnatural) and whether they can be produced by inherent pathways of microorganisms (inherent, noninherent, or created): natural-inherent, natural-noninherent, non-natural-noninherent, and non-natural-created ones. General strategies for systems metabolic engineering of microorganisms for the production of these chemicals using various tools and methods based on omics, genome-scale metabolic modeling and simulation, evolutionary engineering, synthetic biology are suggested with relevant examples. For the production of non-natural chemicals, strategies for the construction of synthetic metabolic pathways are also suggested. Having collected diverse tools and methods for systems metabolic engineering, authors also suggest how to use them and their possible limitations.
Professor Sang Yup Lee said "It is expected that increasing number of chemicals and materials will be produced through biorefineries. We are now equipped with new strategies for developing microbial strains that can produce our desired products at very high efficiencies, thus allowing cost competitiveness to those produced by petrochemical refineries."
Editor of Nature Chemical Biology, Dr. Catherine Goodman, said "It is exciting to see how quickly science is progressing in this field – ideas that used to be science fiction are taking shape in research labs and biorefineries. The article by Professor Lee and his colleagues not only highlights the most advanced techniques and strategies available, but offers critical advice to progress the field as a whole."
The works of Professor Lee have been supported by the Advanced Biomass Center and Intelligent Synthetic Biology Center of Global Frontier Program from the Korean Ministry of Education, Science and Technology through National Research Foundation.
Contact: Dr. Sang Yup Lee, Distinguished Professor and Dean, KAIST, Daejeon, Korea (leesy@kaist.ac.kr, +82-42-350-3930)
2012.05.23 View 16069 -
 Paving the Way to Next Generation Display
A new type of LCD that does not require polymer orientation films has been developed by researchers within the country. This technology will enable the creation of thiner and higher definition display.
Prof. Hee Tae Jung form KAIST’s biochemical engineering department led the research and Hyun Soo Jung, Hwan Jin Jeon doctoral students (1st co-authors), Doctor Yun Ho Kim from Korea Chemistry Research Center, and Prof. Shin Woong Kang from Jeon Buk University ( co-author) have participated in this research. This research has been funded by the WCU program and middle-grade researcher support program. The results of the research has been published as the online update of ‘‘Nature Asia Materials(NPG Asia Materials)” which is a sister magazine of the world renowned academic magazine ‘Nature’.
The flat display industry is the core industry leading the 21st century’s IT industry. The LCD is the main area of research. Korea is the leader of this industry, holding more than 50% of the world market. Many technologies are combined to make the electro-optic devices of the LCD function. The most important technology, which determines the indicating element’s quality and function is the technology to align the liquid crystals in one direction.
Currently, all LCD products are created by mechanically cutting into the surface of the polymer film and orienting the liquid crystal material along these cuts. However, the creation of polymer orientation films cost much time and money, and the high temperature processes necessary to stabilize the polymers does not allow for the free selection of circuit boards, and thus does not allow for the use in flexible display.
Prof. Hee Tae Jung devised a method to orient liquid display without the use of a polymer film using ITOs. Prof. Jung’s base technology has been tested on ITOs to maintain the necessary transparency and conductivity after forming a pattern with high decomposition rates and slenderness ratios.
The technology developed by the research team can horizontally or vertically align the transparent conductors without the use of polymer orientation films. Thus, the manufacturing processes have become much shortened and the LCDs can be made in much thinner from a few micrometers to a few centimeters. Also, it has a lower functioning voltage and faster response speed, showing the prospects of a high definition ultra-fast screen display development. Furthermore, this technology can be used for any type of board, and can be adjusted to a nanometer scale. This enables for its use in LCD based flexible or multi-domain modes.
Also, the transparent conductor patterning technology devised by the research team can be used not only for displays, but also for touch panels with highly increased sensitivity.
Prof. Jung said, “It was a long desire of the industry and academia to find a way to replace the polymer orientation film. This new technology does not need any polymer orientation films, and we can still use the original boards used for LCDs. This mean a lot to the industry. Also, this technology will increase the sensitivity of the touch panels for tablet PCs and smart phones. It can be used in many areas of future electronics base technology.”
2012.04.04 View 12413
Paving the Way to Next Generation Display
A new type of LCD that does not require polymer orientation films has been developed by researchers within the country. This technology will enable the creation of thiner and higher definition display.
Prof. Hee Tae Jung form KAIST’s biochemical engineering department led the research and Hyun Soo Jung, Hwan Jin Jeon doctoral students (1st co-authors), Doctor Yun Ho Kim from Korea Chemistry Research Center, and Prof. Shin Woong Kang from Jeon Buk University ( co-author) have participated in this research. This research has been funded by the WCU program and middle-grade researcher support program. The results of the research has been published as the online update of ‘‘Nature Asia Materials(NPG Asia Materials)” which is a sister magazine of the world renowned academic magazine ‘Nature’.
The flat display industry is the core industry leading the 21st century’s IT industry. The LCD is the main area of research. Korea is the leader of this industry, holding more than 50% of the world market. Many technologies are combined to make the electro-optic devices of the LCD function. The most important technology, which determines the indicating element’s quality and function is the technology to align the liquid crystals in one direction.
Currently, all LCD products are created by mechanically cutting into the surface of the polymer film and orienting the liquid crystal material along these cuts. However, the creation of polymer orientation films cost much time and money, and the high temperature processes necessary to stabilize the polymers does not allow for the free selection of circuit boards, and thus does not allow for the use in flexible display.
Prof. Hee Tae Jung devised a method to orient liquid display without the use of a polymer film using ITOs. Prof. Jung’s base technology has been tested on ITOs to maintain the necessary transparency and conductivity after forming a pattern with high decomposition rates and slenderness ratios.
The technology developed by the research team can horizontally or vertically align the transparent conductors without the use of polymer orientation films. Thus, the manufacturing processes have become much shortened and the LCDs can be made in much thinner from a few micrometers to a few centimeters. Also, it has a lower functioning voltage and faster response speed, showing the prospects of a high definition ultra-fast screen display development. Furthermore, this technology can be used for any type of board, and can be adjusted to a nanometer scale. This enables for its use in LCD based flexible or multi-domain modes.
Also, the transparent conductor patterning technology devised by the research team can be used not only for displays, but also for touch panels with highly increased sensitivity.
Prof. Jung said, “It was a long desire of the industry and academia to find a way to replace the polymer orientation film. This new technology does not need any polymer orientation films, and we can still use the original boards used for LCDs. This mean a lot to the industry. Also, this technology will increase the sensitivity of the touch panels for tablet PCs and smart phones. It can be used in many areas of future electronics base technology.”
2012.04.04 View 12413 -
 Closer to the Dream: Graphene
A technique that allows easy and larger observation area of graphene’s crystal face was developed by Korean Research Team.
The research team, led by Professor Jeong Hui Tae (KAIST), consists of Doctorate candidate Kim Dae Woo, Dr. Kim Yoon Ho (primary author), Doctorate candidate Jeong Hyun Soo. The research is supported by WCU (World Class Research University) Development Plan, Mid-Aged Researcher Support Business and was published in the online edition of Nature Nanotechnology.
(Dissertation: Direct visualization of large0area graphene domains and boundaries by optical birefringency)
Professor Jeong’s team used the optical property of the liquid display used in LCD to visualize the size and shape of the single crystals along a flat surface. The visualization of the single crystal allowed the measurement of a near theoretical value of electrical conductivity of graphene.
Graphene has great electrical conductivity, transparent, mechanically stable, flexible, and is therefore regarded as the next generation electrical material. However the polycrystalinity of graphene meant that the actual electrical, mechanical properties were lower than the theoretical values. The reason was thought to be because of the size of the crystal faces and boundary structures.
Therefore, in order to create graphene that has good properties, observing the domain and boundary of graphene crystal faces is essential.
The new technique developed by the research team is another step towards commercializing transparent electrodes, flexible display, and electric materials like solar cells.
2012.01.31 View 12637
Closer to the Dream: Graphene
A technique that allows easy and larger observation area of graphene’s crystal face was developed by Korean Research Team.
The research team, led by Professor Jeong Hui Tae (KAIST), consists of Doctorate candidate Kim Dae Woo, Dr. Kim Yoon Ho (primary author), Doctorate candidate Jeong Hyun Soo. The research is supported by WCU (World Class Research University) Development Plan, Mid-Aged Researcher Support Business and was published in the online edition of Nature Nanotechnology.
(Dissertation: Direct visualization of large0area graphene domains and boundaries by optical birefringency)
Professor Jeong’s team used the optical property of the liquid display used in LCD to visualize the size and shape of the single crystals along a flat surface. The visualization of the single crystal allowed the measurement of a near theoretical value of electrical conductivity of graphene.
Graphene has great electrical conductivity, transparent, mechanically stable, flexible, and is therefore regarded as the next generation electrical material. However the polycrystalinity of graphene meant that the actual electrical, mechanical properties were lower than the theoretical values. The reason was thought to be because of the size of the crystal faces and boundary structures.
Therefore, in order to create graphene that has good properties, observing the domain and boundary of graphene crystal faces is essential.
The new technique developed by the research team is another step towards commercializing transparent electrodes, flexible display, and electric materials like solar cells.
2012.01.31 View 12637 -
 Future of Petrochemical Industry: The Age of Bio-Refineries
The concept of bio-refinery is based on using biomass from seaweeds and non-edible plant sources to produce various materials.
Bio-refineries has been looked into with increasing interest in modern times due to the advent of global warming (and the subsequent changes in the atmosphere) and the exhaustion of natural resources.
However past 20 years of research in metabolic engineering had a crucial limitation; the need to improve the efficiency of the microorganisms that actually go about converting biomass into biochemical materials.
In order to compensate for the inefficiency, Professor Lee Sang Yeop combined systems biology, composite biology, evolutionary engineering to form ‘systems metabolic engineering’.
This allows combining various data to explain the organism’s state in a multi-dimensional scope and respond accordingly by controlling the metabolism.
The result of the experiment is set as the cover dissertation of ‘Trends in Biotechnology’ magazine’s August edition.
2011.07.28 View 13570
Future of Petrochemical Industry: The Age of Bio-Refineries
The concept of bio-refinery is based on using biomass from seaweeds and non-edible plant sources to produce various materials.
Bio-refineries has been looked into with increasing interest in modern times due to the advent of global warming (and the subsequent changes in the atmosphere) and the exhaustion of natural resources.
However past 20 years of research in metabolic engineering had a crucial limitation; the need to improve the efficiency of the microorganisms that actually go about converting biomass into biochemical materials.
In order to compensate for the inefficiency, Professor Lee Sang Yeop combined systems biology, composite biology, evolutionary engineering to form ‘systems metabolic engineering’.
This allows combining various data to explain the organism’s state in a multi-dimensional scope and respond accordingly by controlling the metabolism.
The result of the experiment is set as the cover dissertation of ‘Trends in Biotechnology’ magazine’s August edition.
2011.07.28 View 13570