research
-
 Algorithm Identifies Optimal Pairs for Composing Metal-Organic Frameworks
The integration of metal-organic frameworks (MOFs) and other metal nanoparticles has increasingly led to the creation of new multifunctional materials. Many researchers have integrated MOFs with other classes of materials to produce new structures with synergetic properties.
Despite there being over 70,000 collections of synthesized MOFs that can be used as building blocks, the precise nature of the interaction and the bonding at the interface between the two materials still remains unknown. The question is how to sort out the right matching pairs out of 70,000 MOFs.
An algorithmic study published in Nature Communications by a KAIST research team presents a clue for finding the perfect pairs. The team, led by Professor Ji-Han Kim from the Department of Chemical and Biomolecular Engineering, developed a joint computational and experimental approach to rationally design MOF@MOFs, a composite of MOFs where an MOF is grown on a different MOF.
Professor Kim’s team, in collaboration with UNIST, noted that the metal node of one MOF can coordinately bond with the linker of a different MOF and the precisely matched interface configurations at atomic and molecular levels can enhance the likelihood of synthesizing MOF@MOFs.
They screened thousands of MOFs and identified optimal MOF pairs that can seamlessly connect to one another by taking advantage of the fact that the metal node of one MOF can form coordination bonds with the linkers of the second MOF. Six pairs predicted from the computational algorithm successfully grew into single crystals.
This computational workflow can readily extend into other classes of materials and can lead to the rapid exploration of the composite MOFs arena for accelerated materials development. Even more, the workflow can enhance the likelihood of synthesizing MOF@MOFs in the form of large single crystals, and thereby demonstrated the utility of rationally designing the MOF@MOFs.
This study is the first algorithm for predicting the synthesis of composite MOFs, to the best of their knowledge. Professor Kim said, “The number of predicted pairs can increase even more with the more general 2D lattice matching, and it is worth investigating in the future.”
This study was supported by Samsung Research Funding & Incubation Center of Samsung Electronics.
(Figure: An example of a rationally synthesized MOF@MOFs (cubic HKUST-1@MOF-5 ))
2019.08.30 View 17500
Algorithm Identifies Optimal Pairs for Composing Metal-Organic Frameworks
The integration of metal-organic frameworks (MOFs) and other metal nanoparticles has increasingly led to the creation of new multifunctional materials. Many researchers have integrated MOFs with other classes of materials to produce new structures with synergetic properties.
Despite there being over 70,000 collections of synthesized MOFs that can be used as building blocks, the precise nature of the interaction and the bonding at the interface between the two materials still remains unknown. The question is how to sort out the right matching pairs out of 70,000 MOFs.
An algorithmic study published in Nature Communications by a KAIST research team presents a clue for finding the perfect pairs. The team, led by Professor Ji-Han Kim from the Department of Chemical and Biomolecular Engineering, developed a joint computational and experimental approach to rationally design MOF@MOFs, a composite of MOFs where an MOF is grown on a different MOF.
Professor Kim’s team, in collaboration with UNIST, noted that the metal node of one MOF can coordinately bond with the linker of a different MOF and the precisely matched interface configurations at atomic and molecular levels can enhance the likelihood of synthesizing MOF@MOFs.
They screened thousands of MOFs and identified optimal MOF pairs that can seamlessly connect to one another by taking advantage of the fact that the metal node of one MOF can form coordination bonds with the linkers of the second MOF. Six pairs predicted from the computational algorithm successfully grew into single crystals.
This computational workflow can readily extend into other classes of materials and can lead to the rapid exploration of the composite MOFs arena for accelerated materials development. Even more, the workflow can enhance the likelihood of synthesizing MOF@MOFs in the form of large single crystals, and thereby demonstrated the utility of rationally designing the MOF@MOFs.
This study is the first algorithm for predicting the synthesis of composite MOFs, to the best of their knowledge. Professor Kim said, “The number of predicted pairs can increase even more with the more general 2D lattice matching, and it is worth investigating in the future.”
This study was supported by Samsung Research Funding & Incubation Center of Samsung Electronics.
(Figure: An example of a rationally synthesized MOF@MOFs (cubic HKUST-1@MOF-5 ))
2019.08.30 View 17500 -
 Researchers Describe a Mechanism Inducing Self-Killing of Cancer Cells
(Professor Kim (left) and lead author Lee)
Researchers have described a new mechanism which induces the self-killing of cancer cells by perturbing ion homeostasis. A research team from the Department of Biochemical Engineering has developed helical polypeptide potassium ionophores that lead to the onset of programmed cell death. The ionophores increase the active oxygen concentration to stress endoplasmic reticulum to the point of cellular death.
The electrochemical gradient between extracellular and intracellular conditions plays an important role in cell growth and metabolism. When a cell’s ion homeostasis is disturbed, critical functions accelerating the activation of apoptosis are inhibited in the cell.
Although ionophores have been intensively used as an ion homeostasis disturber, the mechanisms of cell death have been unclear and the bio-applicability has been limited. In the study featured at Advanced Science, the team presented an alpha helical peptide-based anticancer agent that is capable of transporting potassium ions with water solubility. The cationic, hydrophilic, and potassium ionic groups were combined at the end of the peptide side chain to provide both ion transport and hydrophilic properties.
These peptide-based ionophores reduce the intracellular potassium concentration and at the same time increase the intracellular calcium concentration. Increased intracellular calcium concentrations produce intracellular reactive oxygen species, causing endoplasmic reticulum stress, and ultimately leading to apoptosis.
Anticancer effects were evaluated using tumor-bearing mice to confirm the therapeutic effect, even in animal models. It was found that tumor growth was strongly inhibited by endoplasmic stress-mediated apoptosis.
Lead author Dr. Dae-Yong Lee said, “A peptide-based ionophore is more effective than conventional chemotherapeutic agents because it induces apoptosis via elevated reactive oxygen species levels. Professor Yeu-Chun Kim said he expects this new mechanism to be widely used as a new chemotherapeutic strategy. This research was funded by the National Research Foundation.
2019.08.28 View 21243
Researchers Describe a Mechanism Inducing Self-Killing of Cancer Cells
(Professor Kim (left) and lead author Lee)
Researchers have described a new mechanism which induces the self-killing of cancer cells by perturbing ion homeostasis. A research team from the Department of Biochemical Engineering has developed helical polypeptide potassium ionophores that lead to the onset of programmed cell death. The ionophores increase the active oxygen concentration to stress endoplasmic reticulum to the point of cellular death.
The electrochemical gradient between extracellular and intracellular conditions plays an important role in cell growth and metabolism. When a cell’s ion homeostasis is disturbed, critical functions accelerating the activation of apoptosis are inhibited in the cell.
Although ionophores have been intensively used as an ion homeostasis disturber, the mechanisms of cell death have been unclear and the bio-applicability has been limited. In the study featured at Advanced Science, the team presented an alpha helical peptide-based anticancer agent that is capable of transporting potassium ions with water solubility. The cationic, hydrophilic, and potassium ionic groups were combined at the end of the peptide side chain to provide both ion transport and hydrophilic properties.
These peptide-based ionophores reduce the intracellular potassium concentration and at the same time increase the intracellular calcium concentration. Increased intracellular calcium concentrations produce intracellular reactive oxygen species, causing endoplasmic reticulum stress, and ultimately leading to apoptosis.
Anticancer effects were evaluated using tumor-bearing mice to confirm the therapeutic effect, even in animal models. It was found that tumor growth was strongly inhibited by endoplasmic stress-mediated apoptosis.
Lead author Dr. Dae-Yong Lee said, “A peptide-based ionophore is more effective than conventional chemotherapeutic agents because it induces apoptosis via elevated reactive oxygen species levels. Professor Yeu-Chun Kim said he expects this new mechanism to be widely used as a new chemotherapeutic strategy. This research was funded by the National Research Foundation.
2019.08.28 View 21243 -
 Highly Uniform and Low Hysteresis Pressure Sensor to Increase Practical Applicability
< Professor Steve Park (left) and the First Author Mr. Jinwon Oh (right) >
Researchers have designed a flexible pressure sensor that is expected to have a much wider applicability. A KAIST research team fabricated a piezoresistive pressure sensor of high uniformity with low hysteresis by chemically grafting a conductive polymer onto a porous elastomer template.
The team discovered that the uniformity of pore size and shape is directly related to the uniformity of the sensor. The team noted that by increasing pore size and shape variability, the variability of the sensor characteristics also increases.
Researchers led by Professor Steve Park from the Department of Materials Science and Engineering confirmed that compared to other sensors composed of randomly sized and shaped pores, which had a coefficient of variation in relative resistance change of 69.65%, their newly developed sensor exhibited much higher uniformity with a coefficient of variation of 2.43%. This study was reported in Small as the cover article on August 16.
Flexible pressure sensors have been actively researched and widely applied in electronic equipment such as touch screens, robots, wearable healthcare devices, electronic skin, and human-machine interfaces. In particular, piezoresistive pressure sensors based on elastomer‐conductive material composites hold significant potential due to their many advantages including a simple and low-cost fabrication process.
Various research results have been reported for ways to improve the performance of piezoresistive pressure sensors, most of which have been focused on increasing the sensitivity. Despite its significance, maximizing the sensitivity of composite-based piezoresistive pressure sensors is not necessary for many applications. On the other hand, sensor-to-sensor uniformity and hysteresis are two properties that are of critical importance to realize any application.
The importance of sensor-to-sensor uniformity is obvious. If the sensors manufactured under the same conditions have different properties, measurement reliability is compromised, and therefore the sensor cannot be used in a practical setting.
In addition, low hysteresis is also essential for improved measurement reliability. Hysteresis is a phenomenon in which the electrical readings differ depending on how fast or slow the sensor is being pressed, whether pressure is being released or applied, and how long and to what degree the sensor has been pressed. When a sensor has high hysteresis, the electrical readings will differ even under the same pressure, making the measurements unreliable.
Researchers said they observed a negligible hysteresis degree which was only 2%. This was attributed to the strong chemical bonding between the conductive polymer and the elastomer template, which prevents their relative sliding and displacement, and the porosity of the elastomer that enhances elastic behavior.
“This technology brings forth insight into how to address the two critical issues in pressure sensors: uniformity and hysteresis. We expect our technology to play an important role in increasing practical applications and the commercialization of pressure sensors in the near future,” said Professor Park.
This work was conducted as part of the KAIST‐funded Global Singularity Research Program for 2019, and also supported by the KUSTAR‐KAIST Institute.
Figure 1. Image of a porous elastomer template with uniform pore size and shape (left), Graph showing high uniformity in the sensors’ performance (right).
Figure 2. Hysteresis loops of the sensor at different pressure levels (left), and after a different number of cycles (right).
Figure 3. The cover page of Small Journal, Volume 15, Issue 33.
Publication:
Jinwon Oh, Jin‐Oh Kim, Yunjoo Kim, Han Byul Choi, Jun Chang Yang, Serin Lee, Mikhail Pyatykh, Jung Kim, Joo Yong Sim, and Steve Park. 2019. Highly Uniform and Low Hysteresis Piezoresistive Pressure Sensors Based on Chemical Grafting of Polypyrrole on Elastomer Template with Uniform Pore Size. Small. Wiley-VCH Verlag GmbH & Co. KgaA, Weinheim, Germany, Volume No. 15, Issue No. 33, Full Paper No. 201901744, 8 pages. https://doi.org/10.1002/smll.201901744
Profile: Prof. Steve Park, MS, PhD
stevepark@kaist.ac.kr
http://steveparklab.kaist.ac.kr/
Assistant Professor
Organic and Nano Electronics Laboratory
Department of Materials Science and Engineering
Korea Advanced Institute of Science and Technology (KAIST)
http://kaist.ac.kr
Daejeon 34141, Korea
Profile: Mr. Jinwon Oh, MS
jwoh1701@gmail.com
http://steveparklab.kaist.ac.kr/
Researcher
Organic and Nano Electronics Laboratory
Department of Materials Science and Engineering
Korea Advanced Institute of Science and Technology (KAIST)
http://kaist.ac.kr Daejeon 34141, Korea
Profile: Prof. Jung Kim, MS, PhD
jungkim@kaist.ac.kr
http://medev.kaist.ac.kr/
Professor
Biorobotics Laboratory
Department of Mechanical Engineering
Korea Advanced Institute of Science and Technology (KAIST)
http://kaist.ac.kr Daejeon 34141, Korea
Profile: Joo Yong Sim, PhD
jsim@etri.re.kr
Researcher
Bio-Medical IT Convergence Research Department
Electronics and Telecommunications Research Institute (ETRI)
https://www.etri.re.krDaejeon 34129, Korea
(END)
2019.08.19 View 28827
Highly Uniform and Low Hysteresis Pressure Sensor to Increase Practical Applicability
< Professor Steve Park (left) and the First Author Mr. Jinwon Oh (right) >
Researchers have designed a flexible pressure sensor that is expected to have a much wider applicability. A KAIST research team fabricated a piezoresistive pressure sensor of high uniformity with low hysteresis by chemically grafting a conductive polymer onto a porous elastomer template.
The team discovered that the uniformity of pore size and shape is directly related to the uniformity of the sensor. The team noted that by increasing pore size and shape variability, the variability of the sensor characteristics also increases.
Researchers led by Professor Steve Park from the Department of Materials Science and Engineering confirmed that compared to other sensors composed of randomly sized and shaped pores, which had a coefficient of variation in relative resistance change of 69.65%, their newly developed sensor exhibited much higher uniformity with a coefficient of variation of 2.43%. This study was reported in Small as the cover article on August 16.
Flexible pressure sensors have been actively researched and widely applied in electronic equipment such as touch screens, robots, wearable healthcare devices, electronic skin, and human-machine interfaces. In particular, piezoresistive pressure sensors based on elastomer‐conductive material composites hold significant potential due to their many advantages including a simple and low-cost fabrication process.
Various research results have been reported for ways to improve the performance of piezoresistive pressure sensors, most of which have been focused on increasing the sensitivity. Despite its significance, maximizing the sensitivity of composite-based piezoresistive pressure sensors is not necessary for many applications. On the other hand, sensor-to-sensor uniformity and hysteresis are two properties that are of critical importance to realize any application.
The importance of sensor-to-sensor uniformity is obvious. If the sensors manufactured under the same conditions have different properties, measurement reliability is compromised, and therefore the sensor cannot be used in a practical setting.
In addition, low hysteresis is also essential for improved measurement reliability. Hysteresis is a phenomenon in which the electrical readings differ depending on how fast or slow the sensor is being pressed, whether pressure is being released or applied, and how long and to what degree the sensor has been pressed. When a sensor has high hysteresis, the electrical readings will differ even under the same pressure, making the measurements unreliable.
Researchers said they observed a negligible hysteresis degree which was only 2%. This was attributed to the strong chemical bonding between the conductive polymer and the elastomer template, which prevents their relative sliding and displacement, and the porosity of the elastomer that enhances elastic behavior.
“This technology brings forth insight into how to address the two critical issues in pressure sensors: uniformity and hysteresis. We expect our technology to play an important role in increasing practical applications and the commercialization of pressure sensors in the near future,” said Professor Park.
This work was conducted as part of the KAIST‐funded Global Singularity Research Program for 2019, and also supported by the KUSTAR‐KAIST Institute.
Figure 1. Image of a porous elastomer template with uniform pore size and shape (left), Graph showing high uniformity in the sensors’ performance (right).
Figure 2. Hysteresis loops of the sensor at different pressure levels (left), and after a different number of cycles (right).
Figure 3. The cover page of Small Journal, Volume 15, Issue 33.
Publication:
Jinwon Oh, Jin‐Oh Kim, Yunjoo Kim, Han Byul Choi, Jun Chang Yang, Serin Lee, Mikhail Pyatykh, Jung Kim, Joo Yong Sim, and Steve Park. 2019. Highly Uniform and Low Hysteresis Piezoresistive Pressure Sensors Based on Chemical Grafting of Polypyrrole on Elastomer Template with Uniform Pore Size. Small. Wiley-VCH Verlag GmbH & Co. KgaA, Weinheim, Germany, Volume No. 15, Issue No. 33, Full Paper No. 201901744, 8 pages. https://doi.org/10.1002/smll.201901744
Profile: Prof. Steve Park, MS, PhD
stevepark@kaist.ac.kr
http://steveparklab.kaist.ac.kr/
Assistant Professor
Organic and Nano Electronics Laboratory
Department of Materials Science and Engineering
Korea Advanced Institute of Science and Technology (KAIST)
http://kaist.ac.kr
Daejeon 34141, Korea
Profile: Mr. Jinwon Oh, MS
jwoh1701@gmail.com
http://steveparklab.kaist.ac.kr/
Researcher
Organic and Nano Electronics Laboratory
Department of Materials Science and Engineering
Korea Advanced Institute of Science and Technology (KAIST)
http://kaist.ac.kr Daejeon 34141, Korea
Profile: Prof. Jung Kim, MS, PhD
jungkim@kaist.ac.kr
http://medev.kaist.ac.kr/
Professor
Biorobotics Laboratory
Department of Mechanical Engineering
Korea Advanced Institute of Science and Technology (KAIST)
http://kaist.ac.kr Daejeon 34141, Korea
Profile: Joo Yong Sim, PhD
jsim@etri.re.kr
Researcher
Bio-Medical IT Convergence Research Department
Electronics and Telecommunications Research Institute (ETRI)
https://www.etri.re.krDaejeon 34129, Korea
(END)
2019.08.19 View 28827 -
 Accurate Detection of Low-Level Somatic Mutation in Intractable Epilepsy
KAIST medical scientists have developed an advanced method for perfectly detecting low-level somatic mutation in patients with intractable epilepsy. Their study showed that deep sequencing replicates of major focal epilepsy genes accurately and efficiently identified low-level somatic mutations in intractable epilepsy.
According to the study, their diagnostic method could increase the accuracy up to 100%, unlike the conventional sequencing analysis, which stands at about 30% accuracy. This work was published in Acta Neuropathologica.
Epilepsy is a neurological disorder common in children. Approximately one third of child patients are diagnosed with intractable epilepsy despite adequate anti-epileptic medication treatment.
Somatic mutations in mTOR pathway genes, SLC35A2, and BRAF are the major genetic causes of intractable epilepsies. A clinical trial to target Focal Cortical Dysplasia type II (FCDII), the mTOR inhibitor is underway at Severance Hospital, their collaborator in Seoul, Korea. However, it is difficult to detect such somatic mutations causing intractable epilepsy because their mutational burden is less than 5%, which is similar to the level of sequencing artifacts. In the clinical field, this has remained a standing challenge for the genetic diagnosis of somatic mutations in intractable epilepsy.
Professor Jeong Ho Lee’s team at the Graduate School of Medical Science and Engineering analyzed paired brain and peripheral tissues from 232 intractable epilepsy patients with various brain pathologies at Severance Hospital using deep sequencing and extracted the major focal epilepsy genes.
They narrowed down target genes to eight major focal epilepsy genes, eliminating almost all of the false positive calls using deep targeted sequencing. As a result, the advanced method robustly increased the accuracy and enabled them to detect low-level somatic mutations in unmatched Formalin Fixed Paraffin Embedded (FFPE) brain samples, the most clinically relevant samples.
Professor Lee conducted this study in collaboration with Professor Dong Suk Kim and Hoon-Chul Kang at Severance Hospital of Yonsei University. He said, “This advanced method of genetic analysis will improve overall patient care by providing more comprehensive genetic counseling and informing decisions on alternative treatments.”
Professor Lee has investigated low-level somatic mutations arising in the brain for a decade. He is developing innovative diagnostics and therapeutics for untreatable brain disorders including intractable epilepsy and glioblastoma at a tech-startup called SoVarGen. “All of the technologies we used during the research were transferred to the company. This research gave us very good momentum to reach the next phase of our startup,” he remarked.
The work was supported by grants from the Suh Kyungbae Foundation, a National Research Foundation of Korea grant funded by the Ministry of Science and ICT, the Korean Health Technology R&D Project from the Ministry of Health & Welfare, and the Netherlands Organization for Health Research and Development.
(Figure: Landscape of somatic and germline mutations identified in intractable epilepsy patients. a Signaling pathways for all of the mutated genes identified in this study. Bold: somatic mutation, Regular: germline mutation. b The distribution of variant allelic frequencies (VAFs) of identified somatic mutations. c The detecting rate and types of identified mutations according to histopathology. Yellow: somatic mutations, green: two-hit mutations, grey: germline mutations.)
2019.08.14 View 29705
Accurate Detection of Low-Level Somatic Mutation in Intractable Epilepsy
KAIST medical scientists have developed an advanced method for perfectly detecting low-level somatic mutation in patients with intractable epilepsy. Their study showed that deep sequencing replicates of major focal epilepsy genes accurately and efficiently identified low-level somatic mutations in intractable epilepsy.
According to the study, their diagnostic method could increase the accuracy up to 100%, unlike the conventional sequencing analysis, which stands at about 30% accuracy. This work was published in Acta Neuropathologica.
Epilepsy is a neurological disorder common in children. Approximately one third of child patients are diagnosed with intractable epilepsy despite adequate anti-epileptic medication treatment.
Somatic mutations in mTOR pathway genes, SLC35A2, and BRAF are the major genetic causes of intractable epilepsies. A clinical trial to target Focal Cortical Dysplasia type II (FCDII), the mTOR inhibitor is underway at Severance Hospital, their collaborator in Seoul, Korea. However, it is difficult to detect such somatic mutations causing intractable epilepsy because their mutational burden is less than 5%, which is similar to the level of sequencing artifacts. In the clinical field, this has remained a standing challenge for the genetic diagnosis of somatic mutations in intractable epilepsy.
Professor Jeong Ho Lee’s team at the Graduate School of Medical Science and Engineering analyzed paired brain and peripheral tissues from 232 intractable epilepsy patients with various brain pathologies at Severance Hospital using deep sequencing and extracted the major focal epilepsy genes.
They narrowed down target genes to eight major focal epilepsy genes, eliminating almost all of the false positive calls using deep targeted sequencing. As a result, the advanced method robustly increased the accuracy and enabled them to detect low-level somatic mutations in unmatched Formalin Fixed Paraffin Embedded (FFPE) brain samples, the most clinically relevant samples.
Professor Lee conducted this study in collaboration with Professor Dong Suk Kim and Hoon-Chul Kang at Severance Hospital of Yonsei University. He said, “This advanced method of genetic analysis will improve overall patient care by providing more comprehensive genetic counseling and informing decisions on alternative treatments.”
Professor Lee has investigated low-level somatic mutations arising in the brain for a decade. He is developing innovative diagnostics and therapeutics for untreatable brain disorders including intractable epilepsy and glioblastoma at a tech-startup called SoVarGen. “All of the technologies we used during the research were transferred to the company. This research gave us very good momentum to reach the next phase of our startup,” he remarked.
The work was supported by grants from the Suh Kyungbae Foundation, a National Research Foundation of Korea grant funded by the Ministry of Science and ICT, the Korean Health Technology R&D Project from the Ministry of Health & Welfare, and the Netherlands Organization for Health Research and Development.
(Figure: Landscape of somatic and germline mutations identified in intractable epilepsy patients. a Signaling pathways for all of the mutated genes identified in this study. Bold: somatic mutation, Regular: germline mutation. b The distribution of variant allelic frequencies (VAFs) of identified somatic mutations. c The detecting rate and types of identified mutations according to histopathology. Yellow: somatic mutations, green: two-hit mutations, grey: germline mutations.)
2019.08.14 View 29705 -
 Professor Sang Gyu Kim Receives Yeochon Award for Ecology
Professor Sang-Gyu Kim from the Department of Biological Sciences was selected as the winner of the 12th Yeochon Award for Ecology presented by the Yeochon Association for Ecological Research.
The award was conferred on August 13 in Jeju at the annual conference co-hosted by the Ecological Society of Korea and the Yeochon Association for Ecological Research. Professor Kim received 10 million KRW in prize money.
Professor Kim was recognized for his achievements and contributions in studying herbivorous insects ‘rice weevils’ and their host plant ‘wild tobacco’, especially for having explored the known facts in traditional ecology at the molecular level. His findings are presented in his paper titled ‘Trichobaris weevils distinguish amongst toxic host plants by sensing volatiles that do not affect larval performance’ published in Molecular Ecology in July 2016.
Furthermore, Professor Kim’s research team is continuing their work to identify the ecological functions of plant metabolites as well as interactions between flowers and insect vectors at the molecular level. In doing so, the team edits genes in various plant species using the latest gene editing technology.
The Yeochon Award for Ecology was first established in 2005 with funds donated by a senior ecologist, the late Honorary Professor Joon-Ho Kim of Seoul National University. The award is named after the professor’s pen name “Yeochon” and is intended to encourage promising next-generation ecologists to produce outstanding research achievements in the field of basic ecology.
Professor Kim said, “I will take this award as encouragement to continue taking challenging risks to observe ecological phenomenon from a new perspective. I will continue my research with my students with joy and enthusiasm.”
2019.08.14 View 7417
Professor Sang Gyu Kim Receives Yeochon Award for Ecology
Professor Sang-Gyu Kim from the Department of Biological Sciences was selected as the winner of the 12th Yeochon Award for Ecology presented by the Yeochon Association for Ecological Research.
The award was conferred on August 13 in Jeju at the annual conference co-hosted by the Ecological Society of Korea and the Yeochon Association for Ecological Research. Professor Kim received 10 million KRW in prize money.
Professor Kim was recognized for his achievements and contributions in studying herbivorous insects ‘rice weevils’ and their host plant ‘wild tobacco’, especially for having explored the known facts in traditional ecology at the molecular level. His findings are presented in his paper titled ‘Trichobaris weevils distinguish amongst toxic host plants by sensing volatiles that do not affect larval performance’ published in Molecular Ecology in July 2016.
Furthermore, Professor Kim’s research team is continuing their work to identify the ecological functions of plant metabolites as well as interactions between flowers and insect vectors at the molecular level. In doing so, the team edits genes in various plant species using the latest gene editing technology.
The Yeochon Award for Ecology was first established in 2005 with funds donated by a senior ecologist, the late Honorary Professor Joon-Ho Kim of Seoul National University. The award is named after the professor’s pen name “Yeochon” and is intended to encourage promising next-generation ecologists to produce outstanding research achievements in the field of basic ecology.
Professor Kim said, “I will take this award as encouragement to continue taking challenging risks to observe ecological phenomenon from a new perspective. I will continue my research with my students with joy and enthusiasm.”
2019.08.14 View 7417 -
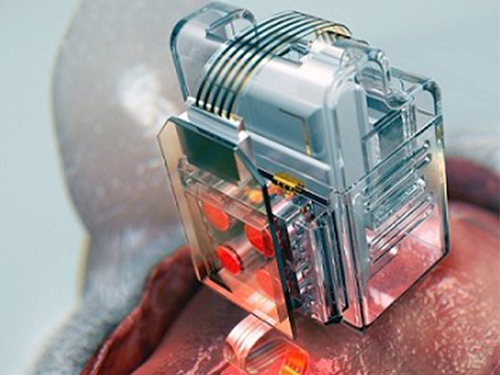 Manipulating Brain Cells by Smartphone
Researchers have developed a soft neural implant that can be wirelessly controlled using a smartphone. It is the first wireless neural device capable of indefinitely delivering multiple drugs and multiple colour lights, which neuroscientists believe can speed up efforts to uncover brain diseases such as Parkinson’s, Alzheimer’s, addiction, depression, and pain.
A team under Professor Jae-Woong Jeong from the School of Electrical Engineering at KAIST and his collaborators have invented a device that can control neural circuits using a tiny brain implant controlled by a smartphone. The device, using Lego-like replaceable drug cartridges and powerful, low-energy Bluetooth, can target specific neurons of interest using drugs and light for prolonged periods. This study was published in Nature Biomedical Engineering.
“This novel device is the fruit of advanced electronics design and powerful micro and nanoscale engineering,” explained Professor Jeong. “We are interested in further developing this technology to make a brain implant for clinical applications.”
This technology significantly overshadows the conventional methods used by neuroscientists, which usually involve rigid metal tubes and optical fibers to deliver drugs and light. Apart from limiting the subject’s movement due to bulky equipment, their relatively rigid structure causes lesions in soft brain tissue over time, therefore making them not suitable for long-term implantation. Although some efforts have been made to partly mitigate adverse tissue response by incorporating soft probes and wireless platforms, the previous solutions were limited by their inability to deliver drugs for long periods of time as well as their bulky and complex control setups.
To achieve chronic wireless drug delivery, scientists had to solve the critical challenge of the exhaustion and evaporation of drugs. To combat this, the researchers invented a neural device with a replaceable drug cartridge, which could allow neuroscientists to study the same brain circuits for several months without worrying about running out of drugs.
These ‘plug-n-play’ drug cartridges were assembled into a brain implant for mice with a soft and ultrathin probe (with the thickness of a human hair), which consisted of microfluidic channels and tiny LEDs (smaller than a grain of salt), for unlimited drug doses and light delivery.
Controlled with an elegant and simple user interface on a smartphone, neuroscientists can easily trigger any specific combination or precise sequencing of light and drug delivery in any implanted target animal without the need to be physically inside the laboratory. Using these wireless neural devices, researchers can also easily setup fully automated animal studies where the behaviour of one animal could affect other animals by triggering light and/or drug delivery.
“The wireless neural device enables chronic chemical and optical neuromodulation that has never been achieved before,” said lead author Raza Qazi, a researcher with KAIST and the University of Colorado Boulder.
This work was supported by grants from the National Research Foundation of Korea, US National Institute of Health, National Institute on Drug Abuse, and Mallinckrodt Professorship.
(A neural implant with replaceable drug cartridges and Bluetooth low-energy can target specific neurons .)
(Micro LED controlling using smartphone application)
2019.08.07 View 33424
Manipulating Brain Cells by Smartphone
Researchers have developed a soft neural implant that can be wirelessly controlled using a smartphone. It is the first wireless neural device capable of indefinitely delivering multiple drugs and multiple colour lights, which neuroscientists believe can speed up efforts to uncover brain diseases such as Parkinson’s, Alzheimer’s, addiction, depression, and pain.
A team under Professor Jae-Woong Jeong from the School of Electrical Engineering at KAIST and his collaborators have invented a device that can control neural circuits using a tiny brain implant controlled by a smartphone. The device, using Lego-like replaceable drug cartridges and powerful, low-energy Bluetooth, can target specific neurons of interest using drugs and light for prolonged periods. This study was published in Nature Biomedical Engineering.
“This novel device is the fruit of advanced electronics design and powerful micro and nanoscale engineering,” explained Professor Jeong. “We are interested in further developing this technology to make a brain implant for clinical applications.”
This technology significantly overshadows the conventional methods used by neuroscientists, which usually involve rigid metal tubes and optical fibers to deliver drugs and light. Apart from limiting the subject’s movement due to bulky equipment, their relatively rigid structure causes lesions in soft brain tissue over time, therefore making them not suitable for long-term implantation. Although some efforts have been made to partly mitigate adverse tissue response by incorporating soft probes and wireless platforms, the previous solutions were limited by their inability to deliver drugs for long periods of time as well as their bulky and complex control setups.
To achieve chronic wireless drug delivery, scientists had to solve the critical challenge of the exhaustion and evaporation of drugs. To combat this, the researchers invented a neural device with a replaceable drug cartridge, which could allow neuroscientists to study the same brain circuits for several months without worrying about running out of drugs.
These ‘plug-n-play’ drug cartridges were assembled into a brain implant for mice with a soft and ultrathin probe (with the thickness of a human hair), which consisted of microfluidic channels and tiny LEDs (smaller than a grain of salt), for unlimited drug doses and light delivery.
Controlled with an elegant and simple user interface on a smartphone, neuroscientists can easily trigger any specific combination or precise sequencing of light and drug delivery in any implanted target animal without the need to be physically inside the laboratory. Using these wireless neural devices, researchers can also easily setup fully automated animal studies where the behaviour of one animal could affect other animals by triggering light and/or drug delivery.
“The wireless neural device enables chronic chemical and optical neuromodulation that has never been achieved before,” said lead author Raza Qazi, a researcher with KAIST and the University of Colorado Boulder.
This work was supported by grants from the National Research Foundation of Korea, US National Institute of Health, National Institute on Drug Abuse, and Mallinckrodt Professorship.
(A neural implant with replaceable drug cartridges and Bluetooth low-energy can target specific neurons .)
(Micro LED controlling using smartphone application)
2019.08.07 View 33424 -
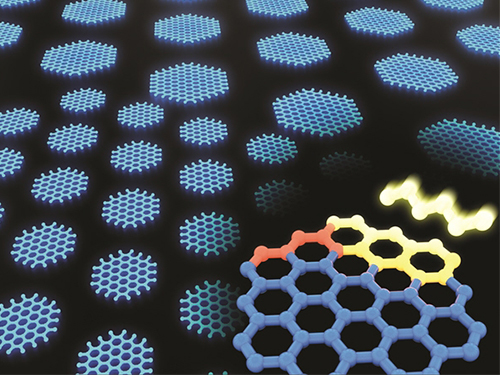 Synthesizing Single-Crystalline Hexagonal Graphene Quantum Dots
(Figure: Uniformly ordered single-crystalline graphene quantum dots of various sizes synthesized through solution chemistry.)
A KAIST team has designed a novel strategy for synthesizing single-crystalline graphene quantum dots, which emit stable blue light. The research team confirmed that a display made of their synthesized graphene quantum dots successfully emitted blue light with stable electric pressure, reportedly resolving the long-standing challenges of blue light emission in manufactured displays. The study, led by Professor O Ok Park in the Department of Chemical and Biological Engineering, was featured online in Nano Letters on July 5.
Graphene has gained increased attention as a next-generation material for its heat and electrical conductivity as well as its transparency. However, single and multi-layered graphene have characteristics of a conductor so that it is difficult to apply into semiconductor. Only when downsized to the nanoscale, semiconductor’s distinct feature of bandgap will be exhibited to emit the light in the graphene. This illuminating featuring of dot is referred to as a graphene quantum dot.
Conventionally, single-crystalline graphene has been fabricated by chemical vapor deposition (CVD) on copper or nickel thin films, or by peeling graphite physically and chemically. However, graphene made via chemical vapor deposition is mainly used for large-surface transparent electrodes. Meanwhile, graphene made by chemical and physical peeling carries uneven size defects.
The research team explained that their graphene quantum dots exhibited a very stable single-phase reaction when they mixed amine and acetic acid with an aqueous solution of glucose. Then, they synthesized single-crystalline graphene quantum dots from the self-assembly of the reaction intermediate. In the course of fabrication, the team developed a new separation method at a low-temperature precipitation, which led to successfully creating a homogeneous nucleation of graphene quantum dots via a single-phase reaction.
Professor Park and his colleagues have developed solution phase synthesis technology that allows for the creation of the desired crystal size for single nanocrystals down to 100 nano meters. It is reportedly the first synthesis of the homogeneous nucleation of graphene through a single-phase reaction.
Professor Park said, "This solution method will significantly contribute to the grafting of graphene in various fields. The application of this new graphene will expand the scope of its applications such as for flexible displays and varistors.”
This research was a joint project with a team from Korea University under Professor Sang Hyuk Im from the Department of Chemical and Biological Engineering, and was supported by the National Research Foundation of Korea, the Nano-Material Technology Development Program from the Electronics and Telecommunications Research Institute (ETRI), KAIST EEWS, and the BK21+ project from the Korean government.
2019.08.02 View 34619
Synthesizing Single-Crystalline Hexagonal Graphene Quantum Dots
(Figure: Uniformly ordered single-crystalline graphene quantum dots of various sizes synthesized through solution chemistry.)
A KAIST team has designed a novel strategy for synthesizing single-crystalline graphene quantum dots, which emit stable blue light. The research team confirmed that a display made of their synthesized graphene quantum dots successfully emitted blue light with stable electric pressure, reportedly resolving the long-standing challenges of blue light emission in manufactured displays. The study, led by Professor O Ok Park in the Department of Chemical and Biological Engineering, was featured online in Nano Letters on July 5.
Graphene has gained increased attention as a next-generation material for its heat and electrical conductivity as well as its transparency. However, single and multi-layered graphene have characteristics of a conductor so that it is difficult to apply into semiconductor. Only when downsized to the nanoscale, semiconductor’s distinct feature of bandgap will be exhibited to emit the light in the graphene. This illuminating featuring of dot is referred to as a graphene quantum dot.
Conventionally, single-crystalline graphene has been fabricated by chemical vapor deposition (CVD) on copper or nickel thin films, or by peeling graphite physically and chemically. However, graphene made via chemical vapor deposition is mainly used for large-surface transparent electrodes. Meanwhile, graphene made by chemical and physical peeling carries uneven size defects.
The research team explained that their graphene quantum dots exhibited a very stable single-phase reaction when they mixed amine and acetic acid with an aqueous solution of glucose. Then, they synthesized single-crystalline graphene quantum dots from the self-assembly of the reaction intermediate. In the course of fabrication, the team developed a new separation method at a low-temperature precipitation, which led to successfully creating a homogeneous nucleation of graphene quantum dots via a single-phase reaction.
Professor Park and his colleagues have developed solution phase synthesis technology that allows for the creation of the desired crystal size for single nanocrystals down to 100 nano meters. It is reportedly the first synthesis of the homogeneous nucleation of graphene through a single-phase reaction.
Professor Park said, "This solution method will significantly contribute to the grafting of graphene in various fields. The application of this new graphene will expand the scope of its applications such as for flexible displays and varistors.”
This research was a joint project with a team from Korea University under Professor Sang Hyuk Im from the Department of Chemical and Biological Engineering, and was supported by the National Research Foundation of Korea, the Nano-Material Technology Development Program from the Electronics and Telecommunications Research Institute (ETRI), KAIST EEWS, and the BK21+ project from the Korean government.
2019.08.02 View 34619 -
 'Flying Drones for Rescue'
(Video Credit: ⓒNASA JPL)
< Team USRG and Professor Shim (second from the right) >
Having recently won the AI R&D Grand Challenge Competition in Korea, Team USRG (Unmanned System Research Group) led by Professor Hyunchul Shim from the School of Electrical Engineering is all geared up to take on their next challenges: the ‘Defense Advanced Research Projects Agency Subterranean Challenge (DARPA SubT Challenge)’ and ‘Lockheed Martin’s AlphaPilot Challenge’ next month.
Team USRG won the obstacle course race in the ‘2019 AI R&D Grand Challenge Competition’ on July 12. They managed to successfully dominate the challenging category of ‘control intelligence.’ Having to complete the obstacle course race solely using AI systems without any connection to the internet made it difficult for most of the eight participating teams to pass the third section of the race, and only Team USRG passed the long pipeline course during their attempt in the main event. They also demonstrated, after the main event, that their drone can navigate all of the checkpoints including landing on the “H” mark using deep learning.
Their drone flew through polls and pipes, and escaped from windows and mazes against strong winds, amid cheers and groans from the crowd gathered at the Korea Exhibition Center (KINTEX) in Goyang, Korea. The team was awarded three million KRW in prize money, and received a research grant worth six hundred million KRW from the Ministry of Science and ICT (MSIT).
“Being ranked first in the race for which we were never given a chance for a test flight means a lot to our team. Considering that we had no information on the exact size of the course in advance, this is a startling result,” said Professor Shim. “We will carry out further research with this funding, and compete once again with the improved AI and drone technology in the 2020 competition,” he added.
The AI R&D Grand Challenge Competition, which was first started in 2017, has been designed to promote AI research and development and expand its application to addressing high-risk technical challenges with significant socio-economic impact.
This year’s competition presented participants with a task where they had to develop AI software technology for drones to navigate themselves autonomously during complex disaster relief operations such as aid delivery.
Each team participated in one of the four tracks of the competition, and their drones were evaluated based on the criteria for each track. The divisions were broken up into intelligent context-awareness, intelligent character recognition, auditory intelligence, and control intelligence.
Team USRG’s technological prowess has been already well acclaimed among international peer groups. Teamed up with NASA JPL, Caltech, and MIT, they will compete in the subterranean mission during the ‘DARPA SubT Challenge’.
Team CoSTAR, as its name stands for, is working together to build ‘Collaborative SubTerranean Autonomous Resilient Robots.’ Professor Shim emphasized the role KAIST plays in Team CoSTAR as a leader in drone technology.
“I think when our drone technology will be added to our peers’ AI and robotics, Team CoSTAR will bring out unsurpassable synergy in completing the subterrestrial and planetary applications. I would like to follow the footprint of Hubo, the winning champion of the 2015 DARPA Robotics Challenge and even extend it to subterranean exploration,” he said.
These next generation autonomous subsurface explorers are now all optimizing the physical AI robot systems developed by Team CoSTAR. They will test their systems in more realistic field environments August 15 through 22 in Pittsburgh, USA. They have already received funding from DARPA for participating.
Team CoSTAR will compete in three consecutive yearly events starting this year, and the last event, planned for 2021, will put the team to the final test with courses that incorporate diverse challenges from all three events. Two million USD will be awarded to the winner after the final event, with additional prizes of up to 200,000 USD for self-funded teams.
Team USRG also ranked third in the recent Hyundai Motor Company’s ‘Autonomous Vehicle Competition’ and another challenge is on the horizon: Lockheed Martin’s ‘AlphaPilot Challenge’. In this event, the teams will be flying their drones through a series of racing gates, trying to beat the best human pilot. The challenge is hosted by Lockheed Martin, the world’s largest military contractor and the maker of the famed F-22 and F-35 stealth fighters, with the goal of stimulating the development of autonomous drones. Team USRG was selected from out of more than 400 teams from around the world and is preparing for a series of races this fall, beginning from the end of August.
Professor Shim said, “It is not easy to perform in a series of competitions in just a few months, but my students are smart, hardworking, and highly motivated. These events indeed demand a lot, but they really challenge the researchers to come up with technologies that work in the real world. This is the way robotics really should be.”
(END)
2019.07.26 View 13532
'Flying Drones for Rescue'
(Video Credit: ⓒNASA JPL)
< Team USRG and Professor Shim (second from the right) >
Having recently won the AI R&D Grand Challenge Competition in Korea, Team USRG (Unmanned System Research Group) led by Professor Hyunchul Shim from the School of Electrical Engineering is all geared up to take on their next challenges: the ‘Defense Advanced Research Projects Agency Subterranean Challenge (DARPA SubT Challenge)’ and ‘Lockheed Martin’s AlphaPilot Challenge’ next month.
Team USRG won the obstacle course race in the ‘2019 AI R&D Grand Challenge Competition’ on July 12. They managed to successfully dominate the challenging category of ‘control intelligence.’ Having to complete the obstacle course race solely using AI systems without any connection to the internet made it difficult for most of the eight participating teams to pass the third section of the race, and only Team USRG passed the long pipeline course during their attempt in the main event. They also demonstrated, after the main event, that their drone can navigate all of the checkpoints including landing on the “H” mark using deep learning.
Their drone flew through polls and pipes, and escaped from windows and mazes against strong winds, amid cheers and groans from the crowd gathered at the Korea Exhibition Center (KINTEX) in Goyang, Korea. The team was awarded three million KRW in prize money, and received a research grant worth six hundred million KRW from the Ministry of Science and ICT (MSIT).
“Being ranked first in the race for which we were never given a chance for a test flight means a lot to our team. Considering that we had no information on the exact size of the course in advance, this is a startling result,” said Professor Shim. “We will carry out further research with this funding, and compete once again with the improved AI and drone technology in the 2020 competition,” he added.
The AI R&D Grand Challenge Competition, which was first started in 2017, has been designed to promote AI research and development and expand its application to addressing high-risk technical challenges with significant socio-economic impact.
This year’s competition presented participants with a task where they had to develop AI software technology for drones to navigate themselves autonomously during complex disaster relief operations such as aid delivery.
Each team participated in one of the four tracks of the competition, and their drones were evaluated based on the criteria for each track. The divisions were broken up into intelligent context-awareness, intelligent character recognition, auditory intelligence, and control intelligence.
Team USRG’s technological prowess has been already well acclaimed among international peer groups. Teamed up with NASA JPL, Caltech, and MIT, they will compete in the subterranean mission during the ‘DARPA SubT Challenge’.
Team CoSTAR, as its name stands for, is working together to build ‘Collaborative SubTerranean Autonomous Resilient Robots.’ Professor Shim emphasized the role KAIST plays in Team CoSTAR as a leader in drone technology.
“I think when our drone technology will be added to our peers’ AI and robotics, Team CoSTAR will bring out unsurpassable synergy in completing the subterrestrial and planetary applications. I would like to follow the footprint of Hubo, the winning champion of the 2015 DARPA Robotics Challenge and even extend it to subterranean exploration,” he said.
These next generation autonomous subsurface explorers are now all optimizing the physical AI robot systems developed by Team CoSTAR. They will test their systems in more realistic field environments August 15 through 22 in Pittsburgh, USA. They have already received funding from DARPA for participating.
Team CoSTAR will compete in three consecutive yearly events starting this year, and the last event, planned for 2021, will put the team to the final test with courses that incorporate diverse challenges from all three events. Two million USD will be awarded to the winner after the final event, with additional prizes of up to 200,000 USD for self-funded teams.
Team USRG also ranked third in the recent Hyundai Motor Company’s ‘Autonomous Vehicle Competition’ and another challenge is on the horizon: Lockheed Martin’s ‘AlphaPilot Challenge’. In this event, the teams will be flying their drones through a series of racing gates, trying to beat the best human pilot. The challenge is hosted by Lockheed Martin, the world’s largest military contractor and the maker of the famed F-22 and F-35 stealth fighters, with the goal of stimulating the development of autonomous drones. Team USRG was selected from out of more than 400 teams from around the world and is preparing for a series of races this fall, beginning from the end of August.
Professor Shim said, “It is not easy to perform in a series of competitions in just a few months, but my students are smart, hardworking, and highly motivated. These events indeed demand a lot, but they really challenge the researchers to come up with technologies that work in the real world. This is the way robotics really should be.”
(END)
2019.07.26 View 13532 -
 KAIST-Google Partnership for AI Education and Research
Google has agreed to support KAIST students and professors in the fields of AI research and education. President Sung-Chul Shin and Google Korea Country Director John Lee signed the collaboration agreement during a ceremony on July 19 at KAIST.
Under the agreement, Google will fund the Google AI-Focused Research Awards Program, the PhD Fellowship Program, and Student Travel Grants for KAIST. In addition, Google will continue to provide more academic and career building opportunities for students, including Google internship programs.
KAIST and Google has been collaborating for years. Professor Steven Whang at the School of Electrical Engineering and Professor Sung Ju Hwang at the School of Computing won the AI-Focused Award in 2018 and conduct their researches on "Improving Generalization and Reliability of Any Deep Neural Networks" and "Automatic and Acitionable Model Analysis for TFX," respectively. Outstanding PhD students have been recognized through the PhD Fellowship Program. However, this new collaboration agreement will focus on research, academic development, and technological innovation in AI.
Google plans to support research in the fields of deep learning, cloud machine learning, and voice technologies. Google will fund the development of two educational programs based on Google open source technology each year for two years that will be used in the new AI Graduate School opening for the fall semester.
John Lee of Google Korea said, “This partnership lays a solid foundation for deeper collaboration.” President Shin added, “This partnership will not only advance Korea’s global competitiveness in AI-powered industries but also contribute to the global community by nurturing talents in this most extensive discipline.”
2019.07.22 View 9269
KAIST-Google Partnership for AI Education and Research
Google has agreed to support KAIST students and professors in the fields of AI research and education. President Sung-Chul Shin and Google Korea Country Director John Lee signed the collaboration agreement during a ceremony on July 19 at KAIST.
Under the agreement, Google will fund the Google AI-Focused Research Awards Program, the PhD Fellowship Program, and Student Travel Grants for KAIST. In addition, Google will continue to provide more academic and career building opportunities for students, including Google internship programs.
KAIST and Google has been collaborating for years. Professor Steven Whang at the School of Electrical Engineering and Professor Sung Ju Hwang at the School of Computing won the AI-Focused Award in 2018 and conduct their researches on "Improving Generalization and Reliability of Any Deep Neural Networks" and "Automatic and Acitionable Model Analysis for TFX," respectively. Outstanding PhD students have been recognized through the PhD Fellowship Program. However, this new collaboration agreement will focus on research, academic development, and technological innovation in AI.
Google plans to support research in the fields of deep learning, cloud machine learning, and voice technologies. Google will fund the development of two educational programs based on Google open source technology each year for two years that will be used in the new AI Graduate School opening for the fall semester.
John Lee of Google Korea said, “This partnership lays a solid foundation for deeper collaboration.” President Shin added, “This partnership will not only advance Korea’s global competitiveness in AI-powered industries but also contribute to the global community by nurturing talents in this most extensive discipline.”
2019.07.22 View 9269 -
 Mathematical Modeling Makes a Breakthrough for a New CRSD Medication
PhD Candidate Dae Wook Kim (Left) and Professor Jae Kyoung Kim (Right)
- Systems approach reveals photosensitivity and PER2 level as determinants of clock-modulator efficacy -
Mathematicians’ new modeling has identified major sources of interspecies and inter-individual variations in the clinical efficacy of a clock-modulating drug: photosensitivity and PER2 level. This enabled precision medicine for circadian disruption.
A KAIST mathematics research team led by Professor Jae Kyoung Kim, in collaboration with Pfizer, applied a combination of mathematical modeling and simulation tools for circadian rhythms sleep disorders (CRSDs) to analyze the animal data generated by Pfizer. This study was reported in Molecular Systems Biology as the cover article on July 8.
Pharmaceutical companies have conducted extensive studies on animals to determine the candidacy of this new medication. However, the results of animal testing do not always translate to the same effects in human trials. Furthermore, even between humans, efficacy differs across individuals depending on an individual’s genetic and environmental factors, which require different treatment strategies.
To overcome these obstacles, KAIST mathematicians and their collaborators developed adaptive chronotherapeutics to identify precise dosing regimens that could restore normal circadian phase under different conditions.
A circadian rhythm is a 24-hour cycle in the physiological processes of living creatures, including humans. A biological clock in the hypothalamic suprachiasmatic nucleus in the human brain sets the time for various human behaviors such as sleep.
A disruption of the endogenous timekeeping system caused by changes in one’s life pattern leads to advanced or delayed sleep-wake cycle phase and a desynchronization between sleep-wake rhythms, resulting in CRSDs. To restore the normal timing of sleep, timing of the circadian clock could be adjusted pharmacologically.
Pfizer identified PF-670462, which can adjust the timing of circadian clock by inhibiting the core clock kinase of the circadian clock (CK1d/e). However, the efficacy of PF-670462 significantly differs between nocturnal mice and diurnal monkeys, whose sleeping times are opposite.
The research team discovered the source of such interspecies variations in drug response by performing thousands of virtual experiments using a mathematical model, which describes biochemical interactions among clock molecules and PF-670462. The result suggests that the effect of PF-670462 is reduced by light exposure in diurnal primates more than in nocturnal mice. This indicates that the strong counteracting effect of light must be considered in order to effectively regulate the circadian clock of diurnal humans using PF-670462.
Furthermore, the team also found the source of inter-patients variations in drug efficacy using virtual patients whose circadian clocks were disrupted due to various mutations. The degree of perturbation in the endogenous level of the core clock molecule PER2 affects the efficacy.
This explains why the clinical outcomes of clock-modulating drugs are highly variable and certain subtypes are unresponsive to treatment. Furthermore, this points out the limitations of current treatment strategies tailored to only the patient’s sleep and wake time but not to the molecular cause of sleep disorders.
PhD candidate Dae Wook Kim, who is the first author, said that this motivates the team to develop an adaptive chronotherapy, which identifies a personalized optimal dosing time of day by tracking the sleep-wake up time of patients via a wearable device and allows for a precision medicine approach for CRSDs.
Professor Jae Kyoung Kim said, "As a mathematician, I am excited to help enable the advancement of a new drug candidate, which can improve the lives of so many patients. I hope this result promotes more collaborations in this translational research.”
This research was supported by a Pfizer grant to KAIST (G01160179), the Human Frontiers Science Program Organization (RGY0063/2017), and a National Research Foundation (NRF) of Korea Grant (NRF-2016 RICIB 3008468 and NRF-2017-Fostering Core Leaders of the Future Basic Science Program/ Global Ph.D. Fellowship Program).
Figure 1. Interspecies and Inter-patients Variations in PF-670462 Efficacy
Figure 2. Journal Cover Page
Publication:
Dae Wook Kim, Cheng Chang, Xian Chen, Angela C Doran, Francois Gaudreault, Travis Wager, George J DeMarco, and Jae Kyoung Kim. 2019. Systems approach reveals photosensitivity and PER2 level as determinants of clock-modulator efficacy. Molecular Systems Biology. EMBO Press, Heidelberg, Germany, Vol. 15, Issue No. 7, Article, 16 pages. https://doi.org/10.15252/msb.20198838
Profile: Prof. Jae Kyoung Kim, PhD
jaekkim@kaist.ac.kr
http://mathsci.kaist.ac.kr/~jaekkim
Associate Professor
Department of Mathematical Sciences
Korea Advanced Institute of Science and Technology (KAIST)
http://kaist.ac.kr Daejeon 34141, Korea
Profile: Dae Wook Kim, PhD Candidate
0308kdo@kaist.ac.kr
http://mathsci.kaist.ac.kr/~jaekkim
PhD Candidate
Department of Mathematical Sciences
Korea Advanced Institute of Science and Technology (KAIST)
http://kaist.ac.kr Daejeon 34141, Korea
Profile: Dr. Cheng Chang, PhD
cheng.chang@pfizer.com
Associate Director of Clinical Pharmacology
Clinical Pharmacology, Global Product Development
Pfizer
https://www.pfizer.com/ Groton 06340, USA
(END)
2019.07.09 View 36423
Mathematical Modeling Makes a Breakthrough for a New CRSD Medication
PhD Candidate Dae Wook Kim (Left) and Professor Jae Kyoung Kim (Right)
- Systems approach reveals photosensitivity and PER2 level as determinants of clock-modulator efficacy -
Mathematicians’ new modeling has identified major sources of interspecies and inter-individual variations in the clinical efficacy of a clock-modulating drug: photosensitivity and PER2 level. This enabled precision medicine for circadian disruption.
A KAIST mathematics research team led by Professor Jae Kyoung Kim, in collaboration with Pfizer, applied a combination of mathematical modeling and simulation tools for circadian rhythms sleep disorders (CRSDs) to analyze the animal data generated by Pfizer. This study was reported in Molecular Systems Biology as the cover article on July 8.
Pharmaceutical companies have conducted extensive studies on animals to determine the candidacy of this new medication. However, the results of animal testing do not always translate to the same effects in human trials. Furthermore, even between humans, efficacy differs across individuals depending on an individual’s genetic and environmental factors, which require different treatment strategies.
To overcome these obstacles, KAIST mathematicians and their collaborators developed adaptive chronotherapeutics to identify precise dosing regimens that could restore normal circadian phase under different conditions.
A circadian rhythm is a 24-hour cycle in the physiological processes of living creatures, including humans. A biological clock in the hypothalamic suprachiasmatic nucleus in the human brain sets the time for various human behaviors such as sleep.
A disruption of the endogenous timekeeping system caused by changes in one’s life pattern leads to advanced or delayed sleep-wake cycle phase and a desynchronization between sleep-wake rhythms, resulting in CRSDs. To restore the normal timing of sleep, timing of the circadian clock could be adjusted pharmacologically.
Pfizer identified PF-670462, which can adjust the timing of circadian clock by inhibiting the core clock kinase of the circadian clock (CK1d/e). However, the efficacy of PF-670462 significantly differs between nocturnal mice and diurnal monkeys, whose sleeping times are opposite.
The research team discovered the source of such interspecies variations in drug response by performing thousands of virtual experiments using a mathematical model, which describes biochemical interactions among clock molecules and PF-670462. The result suggests that the effect of PF-670462 is reduced by light exposure in diurnal primates more than in nocturnal mice. This indicates that the strong counteracting effect of light must be considered in order to effectively regulate the circadian clock of diurnal humans using PF-670462.
Furthermore, the team also found the source of inter-patients variations in drug efficacy using virtual patients whose circadian clocks were disrupted due to various mutations. The degree of perturbation in the endogenous level of the core clock molecule PER2 affects the efficacy.
This explains why the clinical outcomes of clock-modulating drugs are highly variable and certain subtypes are unresponsive to treatment. Furthermore, this points out the limitations of current treatment strategies tailored to only the patient’s sleep and wake time but not to the molecular cause of sleep disorders.
PhD candidate Dae Wook Kim, who is the first author, said that this motivates the team to develop an adaptive chronotherapy, which identifies a personalized optimal dosing time of day by tracking the sleep-wake up time of patients via a wearable device and allows for a precision medicine approach for CRSDs.
Professor Jae Kyoung Kim said, "As a mathematician, I am excited to help enable the advancement of a new drug candidate, which can improve the lives of so many patients. I hope this result promotes more collaborations in this translational research.”
This research was supported by a Pfizer grant to KAIST (G01160179), the Human Frontiers Science Program Organization (RGY0063/2017), and a National Research Foundation (NRF) of Korea Grant (NRF-2016 RICIB 3008468 and NRF-2017-Fostering Core Leaders of the Future Basic Science Program/ Global Ph.D. Fellowship Program).
Figure 1. Interspecies and Inter-patients Variations in PF-670462 Efficacy
Figure 2. Journal Cover Page
Publication:
Dae Wook Kim, Cheng Chang, Xian Chen, Angela C Doran, Francois Gaudreault, Travis Wager, George J DeMarco, and Jae Kyoung Kim. 2019. Systems approach reveals photosensitivity and PER2 level as determinants of clock-modulator efficacy. Molecular Systems Biology. EMBO Press, Heidelberg, Germany, Vol. 15, Issue No. 7, Article, 16 pages. https://doi.org/10.15252/msb.20198838
Profile: Prof. Jae Kyoung Kim, PhD
jaekkim@kaist.ac.kr
http://mathsci.kaist.ac.kr/~jaekkim
Associate Professor
Department of Mathematical Sciences
Korea Advanced Institute of Science and Technology (KAIST)
http://kaist.ac.kr Daejeon 34141, Korea
Profile: Dae Wook Kim, PhD Candidate
0308kdo@kaist.ac.kr
http://mathsci.kaist.ac.kr/~jaekkim
PhD Candidate
Department of Mathematical Sciences
Korea Advanced Institute of Science and Technology (KAIST)
http://kaist.ac.kr Daejeon 34141, Korea
Profile: Dr. Cheng Chang, PhD
cheng.chang@pfizer.com
Associate Director of Clinical Pharmacology
Clinical Pharmacology, Global Product Development
Pfizer
https://www.pfizer.com/ Groton 06340, USA
(END)
2019.07.09 View 36423 -
 Deep Learning-Powered 'DeepEC' Helps Accurately Understand Enzyme Functions
(Figure: Overall scheme of DeepEC)
A deep learning-powered computational framework, ‘DeepEC,’ will allow the high-quality and high-throughput prediction of enzyme commission numbers, which is essential for the accurate understanding of enzyme functions.
A team of Dr. Jae Yong Ryu, Professor Hyun Uk Kim, and Distinguished Professor Sang Yup Lee at KAIST reported the computational framework powered by deep learning that predicts enzyme commission (EC) numbers with high precision in a high-throughput manner.
DeepEC takes a protein sequence as an input and accurately predicts EC numbers as an output. Enzymes are proteins that catalyze biochemical reactions and EC numbers consisting of four level numbers (i.e., a.b.c.d) indicate biochemical reactions. Thus, the identification of EC numbers is critical for accurately understanding enzyme functions and metabolism.
EC numbers are usually given to a protein sequence encoding an enzyme during a genome annotation procedure. Because of the importance of EC numbers, several EC number prediction tools have been developed, but they have room for further improvement with respect to computation time, precision, coverage, and the total size of the files needed for the EC number prediction.
DeepEC uses three convolutional neural networks (CNNs) as a major engine for the prediction of EC numbers, and also implements homology analysis for EC numbers if the three CNNs do not produce reliable EC numbers for a given protein sequence. DeepEC was developed by using a gold standard dataset covering 1,388,606 protein sequences and 4,669 EC numbers.
In particular, benchmarking studies of DeepEC and five other representative EC number prediction tools showed that DeepEC made the most precise and fastest predictions for EC numbers. DeepEC also required the smallest disk space for implementation, which makes it an ideal third-party software component.
Furthermore, DeepEC was the most sensitive in detecting enzymatic function loss as a result of mutations in domains/binding site residue of protein sequences; in this comparative analysis, all the domains or binding site residue were substituted with L-alanine residue in order to remove the protein function, which is known as the L-alanine scanning method.
This study was published online in the Proceedings of the National Academy of Sciences of the United States of America (PNAS) on June 20, 2019, entitled “Deep learning enables high-quality and high-throughput prediction of enzyme commission numbers.”
“DeepEC can be used as an independent tool and also as a third-party software component in combination with other computational platforms that examine metabolic reactions. DeepEC is freely available online,” said Professor Kim.
Distinguished Professor Lee said, “With DeepEC, it has become possible to process ever-increasing volumes of protein sequence data more efficiently and more accurately.”
This work was supported by the Technology Development Program to Solve Climate Changes on Systems Metabolic Engineering for Biorefineries from the Ministry of Science and ICT through the National Research Foundation of Korea. This work was also funded by the Bio & Medical Technology Development Program of the National Research Foundation of Korea funded by the Korean government, the Ministry of Science and ICT.
Profile:
-Professor Hyun Uk Kim (ehukim@kaist.ac.kr)
https://sites.google.com/view/ehukim
Department of Chemical and Biomolecular Engineering
-Distinguished Professor Sang Yup Lee (leesy@kaist.ac.kr)
Department of Chemical and Biomolecular Engineering
http://mbel.kaist.ac.kr
2019.07.09 View 38734
Deep Learning-Powered 'DeepEC' Helps Accurately Understand Enzyme Functions
(Figure: Overall scheme of DeepEC)
A deep learning-powered computational framework, ‘DeepEC,’ will allow the high-quality and high-throughput prediction of enzyme commission numbers, which is essential for the accurate understanding of enzyme functions.
A team of Dr. Jae Yong Ryu, Professor Hyun Uk Kim, and Distinguished Professor Sang Yup Lee at KAIST reported the computational framework powered by deep learning that predicts enzyme commission (EC) numbers with high precision in a high-throughput manner.
DeepEC takes a protein sequence as an input and accurately predicts EC numbers as an output. Enzymes are proteins that catalyze biochemical reactions and EC numbers consisting of four level numbers (i.e., a.b.c.d) indicate biochemical reactions. Thus, the identification of EC numbers is critical for accurately understanding enzyme functions and metabolism.
EC numbers are usually given to a protein sequence encoding an enzyme during a genome annotation procedure. Because of the importance of EC numbers, several EC number prediction tools have been developed, but they have room for further improvement with respect to computation time, precision, coverage, and the total size of the files needed for the EC number prediction.
DeepEC uses three convolutional neural networks (CNNs) as a major engine for the prediction of EC numbers, and also implements homology analysis for EC numbers if the three CNNs do not produce reliable EC numbers for a given protein sequence. DeepEC was developed by using a gold standard dataset covering 1,388,606 protein sequences and 4,669 EC numbers.
In particular, benchmarking studies of DeepEC and five other representative EC number prediction tools showed that DeepEC made the most precise and fastest predictions for EC numbers. DeepEC also required the smallest disk space for implementation, which makes it an ideal third-party software component.
Furthermore, DeepEC was the most sensitive in detecting enzymatic function loss as a result of mutations in domains/binding site residue of protein sequences; in this comparative analysis, all the domains or binding site residue were substituted with L-alanine residue in order to remove the protein function, which is known as the L-alanine scanning method.
This study was published online in the Proceedings of the National Academy of Sciences of the United States of America (PNAS) on June 20, 2019, entitled “Deep learning enables high-quality and high-throughput prediction of enzyme commission numbers.”
“DeepEC can be used as an independent tool and also as a third-party software component in combination with other computational platforms that examine metabolic reactions. DeepEC is freely available online,” said Professor Kim.
Distinguished Professor Lee said, “With DeepEC, it has become possible to process ever-increasing volumes of protein sequence data more efficiently and more accurately.”
This work was supported by the Technology Development Program to Solve Climate Changes on Systems Metabolic Engineering for Biorefineries from the Ministry of Science and ICT through the National Research Foundation of Korea. This work was also funded by the Bio & Medical Technology Development Program of the National Research Foundation of Korea funded by the Korean government, the Ministry of Science and ICT.
Profile:
-Professor Hyun Uk Kim (ehukim@kaist.ac.kr)
https://sites.google.com/view/ehukim
Department of Chemical and Biomolecular Engineering
-Distinguished Professor Sang Yup Lee (leesy@kaist.ac.kr)
Department of Chemical and Biomolecular Engineering
http://mbel.kaist.ac.kr
2019.07.09 View 38734 -
 Hydrogen-Natural Gas Hydrates Harvested by Natural Gas
A hydrogen-natural gas blend (HNGB) can be a game changer only if it can be stored safely and used as a sustainable clean energy resource. A recent study has suggested a new strategy for stably storing hydrogen, using natural gas as a stabilizer. The research proposed a practical gas phase modulator based synthesis of HNGB without generating chemical waste after dissociation for the immediate service.
The research team of Professor Jae Woo Lee from the Department of Chemical and Biomolecular Engineering in collaboration with the Gwangju Institute of Science and Technology (GIST) demonstrated that the natural gas modulator based synthesis leads to significantly reduced synthesis pressure simultaneously with the formation of hydrogen clusters in the confined nanoporous cages of clathrate hydrates. This approach minimizes the environmental impact and reduces operation costs since clathrate hydrates do not generate any chemical waste in both the synthesis and decomposition processes.
For the efficient storage and transportation of hydrogen, numerous materials have been investigated. Among others, clathrate hydrates offer distinct benefits. Clathrate hydrates are nanoporous inclusion compounds composed of a 3D network of polyhedral cages made of hydrogen-bonded ‘host’ water molecules and captured ‘guest’ gas or liquid molecules.
In this study, the research team used two gases, methane and ethane, which have lower equilibrium conditions compared to hydrogen as thermodynamic stabilizers. As a result, they succeeded in stably storing the hydrogen-natural gas compound in hydrates. According to the composition ratio of methane and ethane, structure I or II hydrates can be formed, both of which can stably store hydrogen-natural gas in low-pressure conditions.
The research team found that two hydrogen molecules are stored in small cages in tuned structure I hydrates, while up to three hydrogen molecules can be stored in both small and large cages in tuned structure II hydrates. Hydrates can store gas up to about 170-times its volume and the natural gas used as thermodynamic stabilizers in this study can also be used as an energy source.
The research team developed technology to produce hydrates from ice, produced hydrogen-natural gas hydrates by substitution, and successfully observed that the tuning phenomenon only occurs when hydrogen is involved in hydrate formation from the start for both structures of hydrates.
They expect that the findings can be applied to not only an energy-efficient gas storage material, but also a smart platform to utilize hydrogen natural gas blends, which can serve as a new alternative energy source with targeted hydrogen contents by designing synthetic pathways of mixed gas hydrates.
The research was published online in Energy Storage Materials on June 6, with the title ‘One-step formation of hydrogen clusters in clathrate hydrates stabilized via natural gas blending’.
Professor Lee said, “HNGB will utilize the existing natural gas infrastructure for transportation, so it is very likely that we can commercialize this hydrate system. We are investigating the kinetic performance through a follow-up strategy to increase the volume of gas storage.
This study was funded by the National Research Foundation of Korea and BK21 plus program.
(Figure1. Schematics showing the storage method for hydrogen in a natural gas hydrate using a substitution method and storage method directly from ice to a hydrogen-natural gas hydrate.)
(Figure 2. Artificially synthesized and dissociated hydrogen-natural gas hydrates. The Raman spectra of tuned sI and sII hydrate showing the hydrogen clusters in each cage.)
2019.06.21 View 42218
Hydrogen-Natural Gas Hydrates Harvested by Natural Gas
A hydrogen-natural gas blend (HNGB) can be a game changer only if it can be stored safely and used as a sustainable clean energy resource. A recent study has suggested a new strategy for stably storing hydrogen, using natural gas as a stabilizer. The research proposed a practical gas phase modulator based synthesis of HNGB without generating chemical waste after dissociation for the immediate service.
The research team of Professor Jae Woo Lee from the Department of Chemical and Biomolecular Engineering in collaboration with the Gwangju Institute of Science and Technology (GIST) demonstrated that the natural gas modulator based synthesis leads to significantly reduced synthesis pressure simultaneously with the formation of hydrogen clusters in the confined nanoporous cages of clathrate hydrates. This approach minimizes the environmental impact and reduces operation costs since clathrate hydrates do not generate any chemical waste in both the synthesis and decomposition processes.
For the efficient storage and transportation of hydrogen, numerous materials have been investigated. Among others, clathrate hydrates offer distinct benefits. Clathrate hydrates are nanoporous inclusion compounds composed of a 3D network of polyhedral cages made of hydrogen-bonded ‘host’ water molecules and captured ‘guest’ gas or liquid molecules.
In this study, the research team used two gases, methane and ethane, which have lower equilibrium conditions compared to hydrogen as thermodynamic stabilizers. As a result, they succeeded in stably storing the hydrogen-natural gas compound in hydrates. According to the composition ratio of methane and ethane, structure I or II hydrates can be formed, both of which can stably store hydrogen-natural gas in low-pressure conditions.
The research team found that two hydrogen molecules are stored in small cages in tuned structure I hydrates, while up to three hydrogen molecules can be stored in both small and large cages in tuned structure II hydrates. Hydrates can store gas up to about 170-times its volume and the natural gas used as thermodynamic stabilizers in this study can also be used as an energy source.
The research team developed technology to produce hydrates from ice, produced hydrogen-natural gas hydrates by substitution, and successfully observed that the tuning phenomenon only occurs when hydrogen is involved in hydrate formation from the start for both structures of hydrates.
They expect that the findings can be applied to not only an energy-efficient gas storage material, but also a smart platform to utilize hydrogen natural gas blends, which can serve as a new alternative energy source with targeted hydrogen contents by designing synthetic pathways of mixed gas hydrates.
The research was published online in Energy Storage Materials on June 6, with the title ‘One-step formation of hydrogen clusters in clathrate hydrates stabilized via natural gas blending’.
Professor Lee said, “HNGB will utilize the existing natural gas infrastructure for transportation, so it is very likely that we can commercialize this hydrate system. We are investigating the kinetic performance through a follow-up strategy to increase the volume of gas storage.
This study was funded by the National Research Foundation of Korea and BK21 plus program.
(Figure1. Schematics showing the storage method for hydrogen in a natural gas hydrate using a substitution method and storage method directly from ice to a hydrogen-natural gas hydrate.)
(Figure 2. Artificially synthesized and dissociated hydrogen-natural gas hydrates. The Raman spectra of tuned sI and sII hydrate showing the hydrogen clusters in each cage.)
2019.06.21 View 42218